3S1A
 
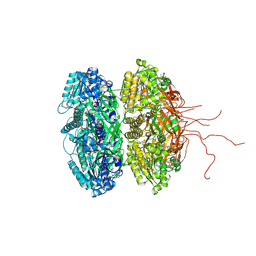 | | Crystal structure of the phosphorylation-site double mutant S431E/T432E of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase kaiC, MAGNESIUM ION | | Authors: | Pattanayek, R, Williams, D.W, Rossi, G, Weigand, S, Mori, T, Johnson, C.H, Stewart, P.L, Egli, M. | | Deposit date: | 2011-05-14 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Combined SAXS/EM Based Models of the S. elongatus Post-Translational Circadian Oscillator and its Interactions with the Output His-Kinase SasA.
Plos One, 6, 2011
|
|
2GBL
 
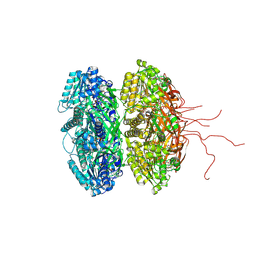 | | Crystal Structure of Full Length Circadian Clock Protein KaiC with Phosphorylation Sites | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase kaiC, MAGNESIUM ION | | Authors: | Pattanayek, R, Williams, D.R, Pattanayek, S, Xu, Y, Mori, T, Johnson, C.H, Stewart, P.L, Egli, M. | | Deposit date: | 2006-03-10 | | Release date: | 2007-01-23 | | Last modified: | 2021-07-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of KaiA-KaiC protein interactions in the cyano-bacterial circadian clock using hybrid structural methods.
Embo J., 25, 2006
|
|
6B20
 
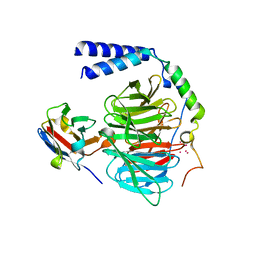 | | Crystal structure of a complex between G protein beta gamma dimer and an inhibitory Nanobody regulator | | Descriptor: | CHLORIDE ION, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, ... | | Authors: | Gulati, S, Kiser, P.D, Palczewski, K. | | Deposit date: | 2017-09-19 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Targeting G protein-coupled receptor signaling at the G protein level with a selective nanobody inhibitor.
Nat Commun, 9, 2018
|
|
2QKE
 
 | |
6OWS
 
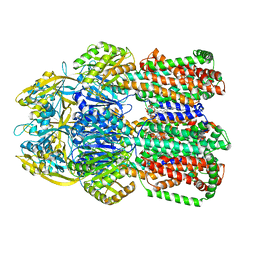 | |
5TE3
 
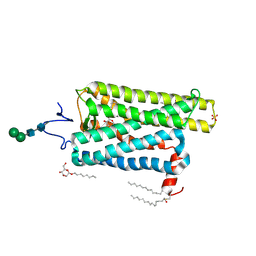 | | Crystal structure of Bos taurus opsin at 2.7 Angstrom | | Descriptor: | PALMITIC ACID, Rhodopsin, SULFATE ION, ... | | Authors: | Gulati, S, Kiser, P.D, Palczewski, K. | | Deposit date: | 2016-09-20 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Photocyclic behavior of rhodopsin induced by an atypical isomerization mechanism.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5TE5
 
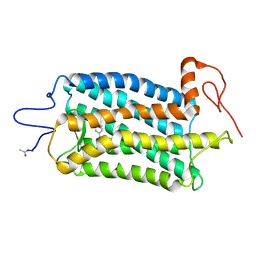 | | Crystal structure of Bos taurus opsin regenerated with 6-carbon ring retinal chromophore | | Descriptor: | (2E)-{(4E)-4-[(3E)-4-(2,6,6-trimethylcyclohex-1-en-1-yl)but-3-en-2-ylidene]cyclohex-2-en-1-ylidene}acetaldehyde, Rhodopsin | | Authors: | Gulati, S, Banerjee, S, Katayama, K, Kiser, P.D, Palczewski, K. | | Deposit date: | 2016-09-20 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.01 Å) | | Cite: | Photocyclic behavior of rhodopsin induced by an atypical isomerization mechanism.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4KSO
 
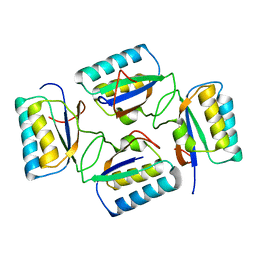 | |
3DVL
 
 | |
7KF8
 
 | |
7KF5
 
 | |
7KF6
 
 | |
7KF7
 
 | |
4HFW
 
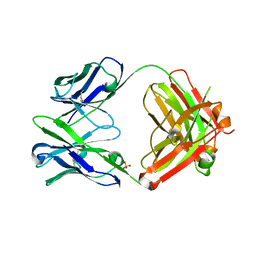 | | Anti Rotavirus Antibody | | Descriptor: | 6-26 Fab Heavy chain, 6-26 Fab Light chain, SULFATE ION | | Authors: | Spiller, B.W, Aiyegbo, M, Crowe, J.E. | | Deposit date: | 2012-10-05 | | Release date: | 2013-05-29 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Human Rotavirus VP6-Specific Antibodies Mediate Intracellular Neutralization by Binding to a Quaternary Structure in the Transcriptional Pore.
Plos One, 8, 2013
|
|
2QZV
 
 | | Draft Crystal Structure of the Vault Shell at 9 Angstroms Resolution | | Descriptor: | Major vault protein | | Authors: | Anderson, D.H, Kickhoefer, V.A, Sievers, S.A, Rome, L.H, Eisenberg, D. | | Deposit date: | 2007-08-17 | | Release date: | 2007-12-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (9 Å) | | Cite: | Draft crystal structure of the vault shell at 9-A resolution.
Plos Biol., 5, 2007
|
|
1CJD
 
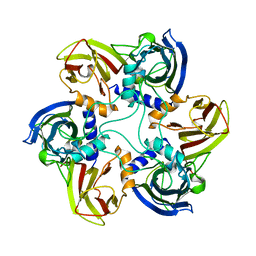 | | THE BACTERIOPHAGE PRD1 COAT PROTEIN, P3, IS STRUCTURALLY SIMILAR TO HUMAN ADENOVIRUS HEXON | | Descriptor: | PROTEIN (MAJOR CAPSID PROTEIN (P3)) | | Authors: | Benson, S.D, Bamford, J.K.H, Bamford, D.H, Burnett, R.M. | | Deposit date: | 1999-04-12 | | Release date: | 1999-09-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Viral evolution revealed by bacteriophage PRD1 and human adenovirus coat protein structures.
Cell(Cambridge,Mass.), 98, 1999
|
|
1HQN
 
 | | THE SELENOMETHIONINE DERIVATIVE OF P3, THE MAJOR COAT PROTEIN OF THE LIPID-CONTAINING BACTERIOPHAGE PRD1. | | Descriptor: | MAJOR CAPSID PROTEIN | | Authors: | Benson, S.D, Bamford, J.K.H, Bamford, D.H, Burnett, R.M. | | Deposit date: | 2000-12-18 | | Release date: | 2001-01-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The X-ray crystal structure of P3, the major coat protein of the lipid-containing bacteriophage PRD1, at 1.65 A resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1HX6
 
 | | P3, THE MAJOR COAT PROTEIN OF THE LIPID-CONTAINING BACTERIOPHAGE PRD1. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, MAJOR CAPSID PROTEIN, ... | | Authors: | Benson, S.D, Bamford, J.K.H, Bamford, D.H, Burnett, R.M. | | Deposit date: | 2001-01-11 | | Release date: | 2001-01-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The X-ray crystal structure of P3, the major coat protein of the lipid-containing bacteriophage PRD1, at 1.65 A resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
