1U7K
 
 | | Structure of a hexameric N-terminal domain from murine leukemia virus capsid | | Descriptor: | Gag polyprotein | | Authors: | Mortuza, G.B, Haire, L.F, Stevens, A, Smerdon, S.J, Stoye, J.P, Taylor, I.A. | | Deposit date: | 2004-08-04 | | Release date: | 2004-10-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | High-resolution structure of a retroviral capsid hexameric amino-terminal domain.
Nature, 431, 2004
|
|
7O2R
 
 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 2 in complex with ITF3985 | | Descriptor: | 3,5-bis(fluoranyl)-~{N}-oxidanyl-4-[(5-pyrimidin-2-yl-1,2,3,4-tetrazol-2-yl)methyl]benzamide, DI(HYDROXYETHYL)ETHER, Histone deacetylase 6, ... | | Authors: | Zrubek, K, Sandrone, G, Cukier, C.D, Stevenazzi, A. | | Deposit date: | 2021-03-31 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of Fluorination in the Histone Deacetylase 6 (HDAC6) Selectivity of Benzohydroxamate-Based Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
7O2P
 
 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 2 in complex with ITF3756 | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zrubek, K, Sandrone, G, Cukier, C.D, Stevenazzi, A. | | Deposit date: | 2021-03-31 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Role of Fluorination in the Histone Deacetylase 6 (HDAC6) Selectivity of Benzohydroxamate-Based Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
8A8Z
 
 | | Crystal structure of Danio rerio HDAC6 CD2 in complex with in situ enzymatically hydrolyzed DFMO-based ITF5924 | | Descriptor: | 4-[[4-[4-(imidazolidin-2-ylideneamino)phenyl]-1,2,3-triazol-1-yl]methyl]benzohydrazide, Histone deacetylase 6, POTASSIUM ION, ... | | Authors: | Zrubek, K, Sandrone, G, Cukier, C.D, Stevenazzi, A. | | Deposit date: | 2022-06-27 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Difluoromethyl-1,3,4-oxadiazoles are slow-binding substrate analog inhibitors of histone deacetylase 6 with unprecedented isotype selectivity.
J.Biol.Chem., 299, 2022
|
|
8FJL
 
 | | Golden Shiner Reovirus Core Tropical Vertex | | Descriptor: | Clamp protein VP6, Major inner capsid protein VP3, Microtubule-associated protein VP5, ... | | Authors: | Stevens, A.S, Zhou, Z.H. | | Deposit date: | 2022-12-19 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Asymmetric reconstruction of the aquareovirus core at near-atomic resolution and mechanism of transcription initiation.
Protein Cell, 14, 2023
|
|
8FJK
 
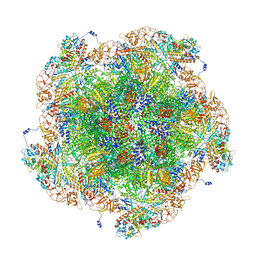 | | Golden Shiner Reovirus Core Polar Vertex | | Descriptor: | Clamp protein VP6, Major inner capsid protein VP3, Microtubule-associated protein VP5, ... | | Authors: | Stevens, A.S, Zhou, Z.H. | | Deposit date: | 2022-12-19 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Asymmetric reconstruction of the aquareovirus core at near-atomic resolution and mechanism of transcription initiation.
Protein Cell, 14, 2023
|
|
7M12
 
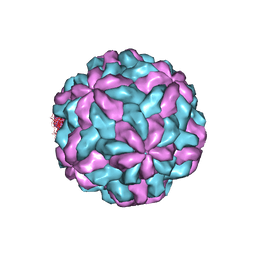 | | TVV2 capsid protein | | Descriptor: | Capsid protein | | Authors: | Zhou, H.Z, Stevens, A.W, Cui, Y.X, Muratore, K.A, Johnson, P.J. | | Deposit date: | 2021-03-12 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Atomic Structure of the Trichomonas vaginalis Double-Stranded RNA Virus 2.
Mbio, 12, 2021
|
|
7LWY
 
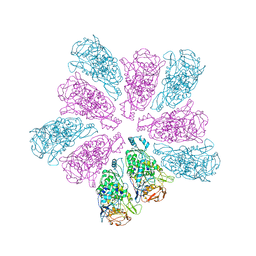 | | TVV viral capsid protein | | Descriptor: | Capsid protein | | Authors: | Zhou, Z.H, Stevens, A.W, Cui, Y.X, Johnson, P.J, Muratore, K.A. | | Deposit date: | 2021-03-02 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Atomic Structure of the Trichomonas vaginalis Double-Stranded RNA Virus 2.
Mbio, 12, 2021
|
|
4D1Q
 
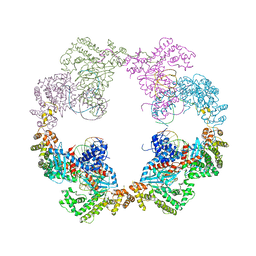 | | Hermes transposase bound to its terminal inverted repeat | | Descriptor: | SODIUM ION, TERMINAL INVERTED REPEAT, TRANSPOSASE | | Authors: | Hickman, A.B, Ewis, H, Li, X, Knapp, J, Laver, T, Doss, A.L, Tolun, G, Steven, A, Grishaev, A, Bax, A, Atkinson, P, Craig, N.L, Dyda, F. | | Deposit date: | 2014-05-04 | | Release date: | 2014-07-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Basis of Hat Transposon End Recognition by Hermes, an Octameric DNA Transposase from Musca Domestica.
Cell(Cambridge,Mass.), 158, 2014
|
|
6UI7
 
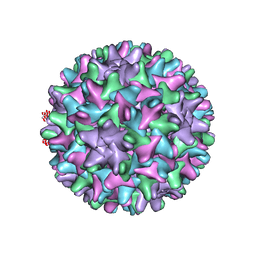 | | HBV T=4 149C3A | | Descriptor: | Core protein | | Authors: | Wu, W, Watts, N.R, Cheng, N, Huang, R, Steven, A, Wingfield, P.T. | | Deposit date: | 2019-09-30 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Expression of quasi-equivalence and capsid dimorphism in the Hepadnaviridae.
Plos Comput.Biol., 16, 2020
|
|
6UI6
 
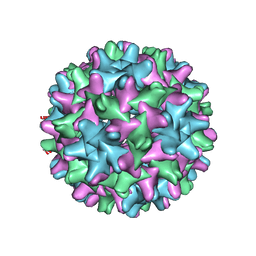 | | HBV T=3 149C3A | | Descriptor: | Core protein | | Authors: | Wu, W, Watts, N.R, Cheng, N, Huang, R, Steven, A, Wingfield, P.T. | | Deposit date: | 2019-09-30 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Expression of quasi-equivalence and capsid dimorphism in the Hepadnaviridae.
Plos Comput.Biol., 16, 2020
|
|
1Q67
 
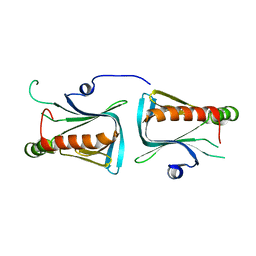 | | Crystal structure of Dcp1p | | Descriptor: | Decapping protein involved in mRNA degradation-Dcp1p | | Authors: | She, M, Decker, C.J, Liu, Y, Chen, N, Parker, R, Song, H. | | Deposit date: | 2003-08-12 | | Release date: | 2004-03-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Dcp1p and its functional implications in mRNA decapping
Nat.Struct.Mol.Biol., 11, 2004
|
|
