1B0T
 
 | | D15K/K84D MUTANT OF AZOTOBACTER VINELANDII FDI | | Descriptor: | FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, PROTEIN (FERREDOXIN I) | | Authors: | Sridhar, V, Stout, C.D, Chen, K, Kemper, M.A, Burgess, B.K. | | Deposit date: | 1998-11-12 | | Release date: | 1998-11-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the D15K/K84D Mutant of Azotobacter Vinelandii Ferredoxin I
To be Published
|
|
4LKQ
 
 | | Crystal structure of PDE10A2 with fragment ZT017 | | Descriptor: | 4-amino-1,7-dihydro-6H-pyrazolo[3,4-d]pyrimidine-6-thione, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-07-08 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Identification and Optimization of PDE10A Inhibitors Using Fragment-Based Screening by Nanocalorimetry and X-ray Crystallography.
J Biomol Screen, 19, 2014
|
|
4LLP
 
 | | Crystal structure of PDE10A2 with fragment ZT401 | | Descriptor: | 4-amino-2-methylphenol, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification and Optimization of PDE10A Inhibitors Using Fragment-Based Screening by Nanocalorimetry and X-ray Crystallography.
J Biomol Screen, 19, 2014
|
|
4LM4
 
 | | Crystal structure of PDE10A2 with fragment ZT902 | | Descriptor: | NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A, quinazolin-4(1H)-one | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-07-10 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Identification and Optimization of PDE10A Inhibitors Using Fragment-Based Screening by Nanocalorimetry and X-ray Crystallography.
J Biomol Screen, 19, 2014
|
|
4LM1
 
 | | Crystal structure of PDE10A2 with fragment ZT450 | | Descriptor: | 5-nitroquinoline, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification and Optimization of PDE10A Inhibitors Using Fragment-Based Screening by Nanocalorimetry and X-ray Crystallography.
J Biomol Screen, 19, 2014
|
|
4LLX
 
 | | Crystal structure of PDE10A2 with fragment ZT434 | | Descriptor: | 4,6-dimethylpyrimidin-2-amine, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification and Optimization of PDE10A Inhibitors Using Fragment-Based Screening by Nanocalorimetry and X-ray Crystallography.
J Biomol Screen, 19, 2014
|
|
4LM3
 
 | | Crystal structure of PDE10A2 with fragment ZT464 | | Descriptor: | 1,2-ETHANEDIOL, 1-(2,3-dihydro-1,4-benzodioxin-6-yl)ethanone, NICKEL (II) ION, ... | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Identification and Optimization of PDE10A Inhibitors Using Fragment-Based Screening by Nanocalorimetry and X-ray Crystallography.
J Biomol Screen, 19, 2014
|
|
4LLJ
 
 | | Crystal structure of PDE10A2 with fragment ZT214 | | Descriptor: | 2H-isoindole-1,3-diamine, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Identification and Optimization of PDE10A Inhibitors Using Fragment-Based Screening by Nanocalorimetry and X-ray Crystallography.
J Biomol Screen, 19, 2014
|
|
4LM2
 
 | | Crystal structure of PDE10A2 with fragment ZT462 | | Descriptor: | 2,3-dihydro-1,4-benzodioxin-6-ylmethanol, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification and Optimization of PDE10A Inhibitors Using Fragment-Based Screening by Nanocalorimetry and X-ray Crystallography.
J Biomol Screen, 19, 2014
|
|
4LM0
 
 | | Crystal structure of PDE10A2 with fragment ZT448 | | Descriptor: | 5-NITROINDAZOLE, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Identification and Optimization of PDE10A Inhibitors Using Fragment-Based Screening by Nanocalorimetry and X-ray Crystallography.
J Biomol Screen, 19, 2014
|
|
4LLK
 
 | | Crystal structure of PDE10A2 with fragment ZT217 | | Descriptor: | 2-methylquinazolin-4(3H)-one, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification and Optimization of PDE10A Inhibitors Using Fragment-Based Screening by Nanocalorimetry and X-ray Crystallography.
J Biomol Screen, 19, 2014
|
|
4MRW
 
 | | Crystal structure of PDE10A2 with fragment ZT0120 (7-chloroquinolin-4-ol) | | Descriptor: | 7-chloroquinolin-4-ol, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, NIenaber, V. | | Deposit date: | 2013-09-17 | | Release date: | 2014-05-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Identification and optimization of PDE10A inhibitors using fragment-based screening by nanocalorimetry and X-ray crystallography.
J Biomol Screen, 19, 2014
|
|
4MSH
 
 | | Crystal Structure of PDE10A2 with fragment ZT0143 ((2S)-4-chloro-2,3-dihydro-1,3-benzothiazol-2-amine) | | Descriptor: | 4-chloro-1,3-benzothiazol-2-amine, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-09-18 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification and optimization of PDE10A inhibitors using fragment-based screening by nanocalorimetry and X-ray crystallography.
J Biomol Screen, 19, 2014
|
|
4MSC
 
 | | Crystal structure of PDE10A2 with fragment ZT1595 (2-[(quinolin-7-yloxy)methyl]quinoline) | | Descriptor: | 2-[(quinolin-7-yloxy)methyl]quinoline, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-09-18 | | Release date: | 2014-05-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Identification and optimization of PDE10A inhibitors using fragment-based screening by nanocalorimetry and X-ray crystallography.
J Biomol Screen, 19, 2014
|
|
4MSE
 
 | | Crystal structure of PDE10A2 with fragment ZT1597 (2-({[(2S)-2-methyl-2,3-dihydro-1,3-benzothiazol-5-yl]oxy}methyl)quinoline) | | Descriptor: | 2-{[(2-methyl-1,3-benzothiazol-5-yl)oxy]methyl}quinoline, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-09-18 | | Release date: | 2014-05-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Identification and optimization of PDE10A inhibitors using fragment-based screening by nanocalorimetry and X-ray crystallography.
J Biomol Screen, 19, 2014
|
|
4MRZ
 
 | | Crystal structure of PDE10A2 with fragment ZT0429 (4-methyl-3-nitropyridin-2-amine) | | Descriptor: | 4-methyl-3-nitropyridin-2-amine, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-09-17 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Identification and optimization of PDE10A inhibitors using fragment-based screening by nanocalorimetry and X-ray crystallography.
J Biomol Screen, 19, 2014
|
|
4MS0
 
 | | Crystal structure of PDE10A2 with fragment ZT0443 (6-chloropyrimidine-2,4-diamine) | | Descriptor: | 6-chloropyrimidine-2,4-diamine, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-09-18 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Identification and optimization of PDE10A inhibitors using fragment-based screening by nanocalorimetry and X-ray crystallography.
J Biomol Screen, 19, 2014
|
|
4MSN
 
 | | Crystal structure of PDE10A2 with fragment ZT0451 (8-nitroquinoline) | | Descriptor: | 8-nitroquinoline, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-09-18 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification and optimization of PDE10A inhibitors using fragment-based screening by nanocalorimetry and X-ray crystallography.
J Biomol Screen, 19, 2014
|
|
1B0V
 
 | | I40N MUTANT OF AZOTOBACTER VINELANDII FDI | | Descriptor: | FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, PROTEIN (FERREDOXIN) | | Authors: | Sridhar, V, Prasad, G.S, Stout, C.D, Chen, K, Burgess, B.K. | | Deposit date: | 1998-11-12 | | Release date: | 2000-01-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Alteration of the reduction potential of the [4Fe-4S](2+/+) cluster of Azotobacter vinelandii ferredoxin I.
J.Biol.Chem., 274, 1999
|
|
4POK
 
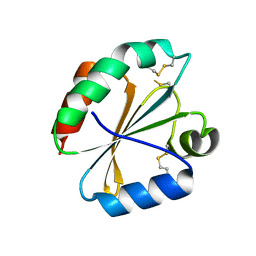 | | Crystal structures of thioredoxin with mesna at 2.5A resolution | | Descriptor: | 1-THIOETHANESULFONIC ACID, Thioredoxin | | Authors: | Sridhar, V, Chie-Leon, B, Badger, J, Nienaber, V.L, Hausheer, F.H. | | Deposit date: | 2014-02-25 | | Release date: | 2014-10-29 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | BNP7787 Forms Novel Covalent
Adducts on Human Thioredoxin
and Modulates Thioredoxin
Activity
J Pharmacol Clin Toxicol, 2, 2014
|
|
4POM
 
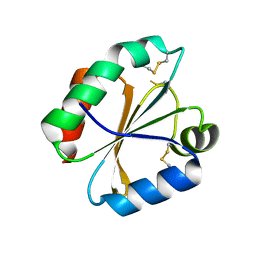 | | Crystal structures of thioredoxin with mesna at 1.85A resolution | | Descriptor: | 1-THIOETHANESULFONIC ACID, Thioredoxin | | Authors: | Sridhar, V, Chie-Leon, B, Badger, J, Nienaber, V.L, Hausheer, F.H. | | Deposit date: | 2014-02-26 | | Release date: | 2014-10-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | BNP7787 Forms Novel Covalent
Adducts on Human Thioredoxin
and Modulates Thioredoxin
Activity
J Pharmacol Clin Toxicol, 2, 2014
|
|
4POL
 
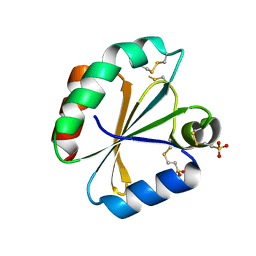 | | Crystal structures of thioredoxin with mesna at 2.8A resolution | | Descriptor: | 1-THIOETHANESULFONIC ACID, Thioredoxin | | Authors: | Sridhar, V, Chie-Leon, B, Badger, J, Nienaber, V.L, Hausheer, F.H. | | Deposit date: | 2014-02-26 | | Release date: | 2014-10-29 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | BNP7787 Forms Novel Covalent
Adducts on Human Thioredoxin
and Modulates Thioredoxin
Activity
J Pharmacol Clin Toxicol, 2, 2014
|
|
1L7E
 
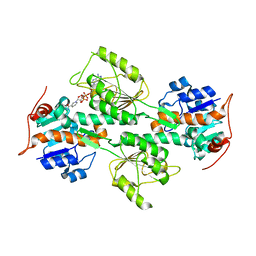 | | Crystal Structure of R. rubrum Transhydrogenase Domain I with Bound NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, nicotinamide nucleotide Transhydrogenase, subunit alpha 1 | | Authors: | Prasad, G.S, Wahlberg, M, Sridhar, V, Yamaguchi, M, Hatefi, Y, Stout, C.D. | | Deposit date: | 2002-03-14 | | Release date: | 2002-11-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Transhydrogenase Domain I
with and without Bound NADH
Biochemistry, 41, 2002
|
|
3PMO
 
 | | The structure of LpxD from Pseudomonas aeruginosa at 1.3 A resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, UDP-3-O-[3-hydroxymyristoyl] glucosamine N-acyltransferase | | Authors: | Badger, J, Chie-Leon, B, Logan, C, Sridhar, V, Sankaran, B, Zwart, P.H, Nienaber, V. | | Deposit date: | 2010-11-17 | | Release date: | 2011-07-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The structure of LpxD from Pseudomonas aeruginosa at 1.3 A resolution.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3M1B
 
 | | Crystal structure of human FcRn with a dimeric peptide inhibitor | | Descriptor: | Beta-2-microglobulin, DIMERIC PEPTIDE INHIBITOR, IgG receptor FcRn large subunit p51 | | Authors: | Mezo, A.R, Sridhar, V, Badger, J, Sakorafas, P, Nienaber, V. | | Deposit date: | 2010-03-04 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | X-ray crystal structures of monomeric and dimeric peptide inhibitors in complex with the human neonatal Fc receptor, FcRn.
J.Biol.Chem., 285, 2010
|
|
