1AVX
 
 | | COMPLEX PORCINE PANCREATIC TRYPSIN/SOYBEAN TRYPSIN INHIBITOR, TETRAGONAL CRYSTAL FORM | | Descriptor: | CALCIUM ION, TRYPSIN, TRYPSIN INHIBITOR | | Authors: | Song, H.K, Suh, S.W. | | Deposit date: | 1997-09-21 | | Release date: | 1998-10-28 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Kunitz-type soybean trypsin inhibitor revisited: refined structure of its complex with porcine trypsin reveals an insight into the interaction between a homologous inhibitor from Erythrina caffra and tissue-type plasminogen activator.
J.Mol.Biol., 275, 1998
|
|
1AVW
 
 | | COMPLEX PORCINE PANCREATIC TRYPSIN/SOYBEAN TRYPSIN INHIBITOR, ORTHORHOMBIC CRYSTAL FORM | | Descriptor: | CALCIUM ION, TRYPSIN, TRYPSIN INHIBITOR | | Authors: | Song, H.K, Suh, S.W. | | Deposit date: | 1997-09-21 | | Release date: | 1998-10-28 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Kunitz-type soybean trypsin inhibitor revisited: refined structure of its complex with porcine trypsin reveals an insight into the interaction between a homologous inhibitor from Erythrina caffra and tissue-type plasminogen activator.
J.Mol.Biol., 275, 1998
|
|
1AVU
 
 | | TRYPSIN INHIBITOR FROM SOYBEAN (STI) | | Descriptor: | TRYPSIN INHIBITOR | | Authors: | Song, H.K, Suh, S.W. | | Deposit date: | 1997-09-20 | | Release date: | 1998-10-28 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Kunitz-type soybean trypsin inhibitor revisited: refined structure of its complex with porcine trypsin reveals an insight into the interaction between a homologous inhibitor from Erythrina caffra and tissue-type plasminogen activator.
J.Mol.Biol., 275, 1998
|
|
1C2A
 
 | | CRYSTAL STRUCTURE OF BARLEY BBI | | Descriptor: | BOWMAN-BIRK TRYPSIN INHIBITOR | | Authors: | Song, H.K, Kim, Y.S, Yang, J.K, Moon, J, Lee, J.Y, Suh, S.W. | | Deposit date: | 1999-07-23 | | Release date: | 1999-12-29 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a 16 kDa double-headed Bowman-Birk trypsin inhibitor from barley seeds at 1.9 A resolution.
J.Mol.Biol., 293, 1999
|
|
1C02
 
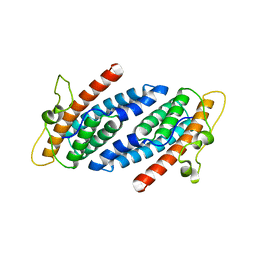 | | CRYSTAL STRUCTURE OF YEAST YPD1P | | Descriptor: | PHOSPHOTRANSFERASE YPD1P | | Authors: | Song, H.K, Lee, J.Y, Lee, M.G, Suh, S.W. | | Deposit date: | 1999-07-14 | | Release date: | 2000-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into eukaryotic multistep phosphorelay signal transduction revealed by the crystal structure of Ypd1p from Saccharomyces cerevisiae.
J.Mol.Biol., 293, 1999
|
|
1C03
 
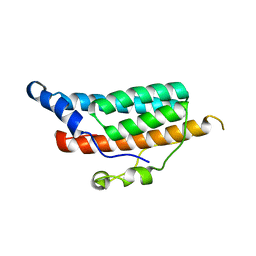 | | CRYSTAL STRUCTURE OF YPD1P (TRICLINIC FORM) | | Descriptor: | HYPOTHETICAL PROTEIN YDL235C | | Authors: | Song, H.K, Lee, J.Y, Lee, M.G, Suh, S.W. | | Deposit date: | 1999-07-14 | | Release date: | 2000-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insights into eukaryotic multistep phosphorelay signal transduction revealed by the crystal structure of Ypd1p from Saccharomyces cerevisiae.
J.Mol.Biol., 293, 1999
|
|
1CNS
 
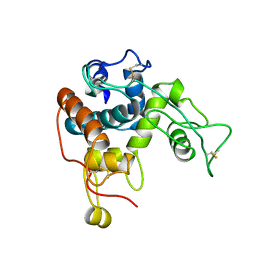 | |
1E94
 
 | | HslV-HslU from E.coli | | Descriptor: | HEAT SHOCK PROTEIN HSLU, HEAT SHOCK PROTEIN HSLV, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Song, H.K, Hartmann, C, Ravishankar, R, Bochtler, M. | | Deposit date: | 2000-10-07 | | Release date: | 2000-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mutational Studies on Hslu and its Docking Mode with Hslv
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1B5E
 
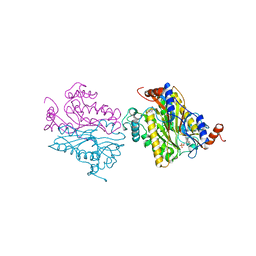 | | DCMP HYDROXYMETHYLASE FROM T4 | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, PROTEIN (DEOXYCYTIDYLATE HYDROXYMETHYLASE) | | Authors: | Song, H.K, Sohn, S.H, Suh, S.W. | | Deposit date: | 1999-01-06 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of deoxycytidylate hydroxymethylase from bacteriophage T4, a component of the deoxyribonucleoside triphosphate-synthesizing complex.
EMBO J., 18, 1999
|
|
1B5D
 
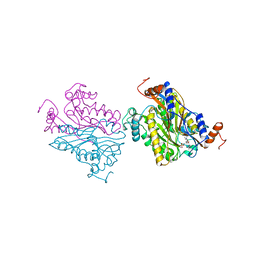 | | DCMP Hydroxymethylase from T4 (Intact) | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, PROTEIN (DEOXYCYTIDYLATE HYDROXYMETHYLASE) | | Authors: | Song, H.K, Sohn, S.H, Suh, S.W. | | Deposit date: | 1999-01-06 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of deoxycytidylate hydroxymethylase from bacteriophage T4, a component of the deoxyribonucleoside triphosphate-synthesizing complex.
EMBO J., 18, 1999
|
|
1B49
 
 | | DCMP HYDROXYMETHYLASE FROM T4 (PHOSPHATE-BOUND) | | Descriptor: | PHOSPHATE ION, PROTEIN (DEOXYCYTIDYLATE HYDROXYMETHYLASE) | | Authors: | Song, H.K, Sohn, S.H, Suh, S.W. | | Deposit date: | 1999-01-06 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of deoxycytidylate hydroxymethylase from bacteriophage T4, a component of the deoxyribonucleoside triphosphate-synthesizing complex.
EMBO J., 18, 1999
|
|
3BSF
 
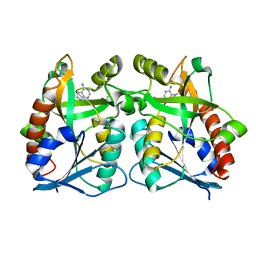 | |
1KCT
 
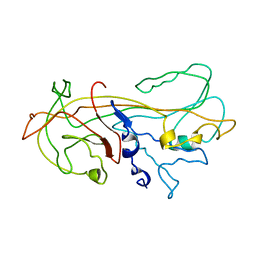 | | ALPHA1-ANTITRYPSIN | | Descriptor: | ALPHA1-ANTITRYPSIN | | Authors: | Song, H.K, Suh, S.W. | | Deposit date: | 1996-08-06 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Crystal structure of an uncleaved alpha 1-antitrypsin reveals the conformation of its inhibitory reactive loop.
FEBS Lett., 377, 1995
|
|
2FJ8
 
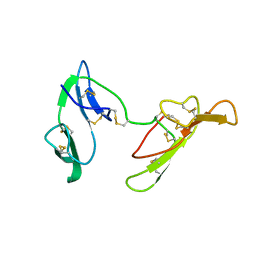 | |
1OX8
 
 | | Crystal structure of SspB | | Descriptor: | Stringent starvation protein B | | Authors: | Song, H.K, Eck, M.J. | | Deposit date: | 2003-04-01 | | Release date: | 2003-08-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of degradation signal recognition by SspB, a specificity-enhancing factor for the ClpXP proteolytic machine
Mol.Cell, 12, 2003
|
|
1M4Y
 
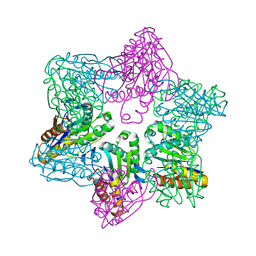 | | Crystal structure of HslV from Thermotoga maritima | | Descriptor: | ATP-dependent protease hslV, SODIUM ION | | Authors: | Song, H.K, Ramachandran, R, Bochtler, M.B, Hartmann, C, Azim, M.K, Huber, R. | | Deposit date: | 2002-07-05 | | Release date: | 2003-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Isolation and characterization of the prokaryotic proteasome homolog HslVU (ClpQY) from Thermotoga maritima and the crystal structure of HslV.
BIOPHYS.CHEM., 100, 2003
|
|
1OX9
 
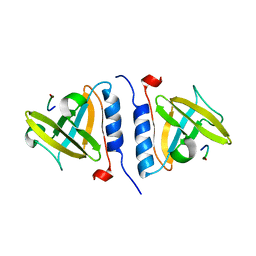 | | Crystal structure of SspB-ssrA complex | | Descriptor: | Stringent starvation protein B, ssrA | | Authors: | Song, H.K, Eck, M.J. | | Deposit date: | 2003-04-01 | | Release date: | 2003-08-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of degradation signal recognition by SspB, a specificity-enhancing factor for the ClpXP proteolytic machine
Mol.Cell, 12, 2003
|
|
1GMW
 
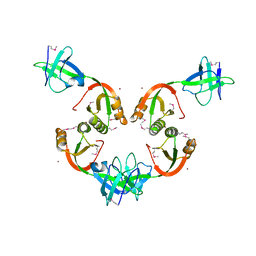 | | Structure of UreE | | Descriptor: | COPPER (II) ION, UREE | | Authors: | Song, H.K, Mulrooney, S.B, Huber, R, Hausinger, R. | | Deposit date: | 2001-09-24 | | Release date: | 2001-11-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Klebsiella Aerogenes Uree, a Nickel-Binding Metallochaperone for Urease Activation
J.Biol.Chem., 276, 2001
|
|
1GMV
 
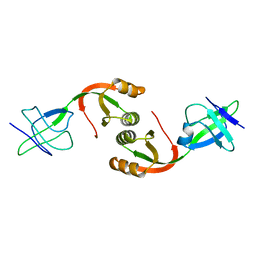 | | Structure of UreE | | Descriptor: | UREE | | Authors: | Song, H.K, Mulrooney, S.B, Huber, R, Hausinger, R. | | Deposit date: | 2001-09-24 | | Release date: | 2001-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Klebsiella Aerogenes Uree, a Nickel-Binding Metallochaperone for Urease Activation.
J.Biol.Chem., 276, 2001
|
|
1GMU
 
 | | Structure of UreE | | Descriptor: | UREE | | Authors: | Song, H.K, Mulrooney, S.B, Huber, R, Hausinger, R. | | Deposit date: | 2001-09-24 | | Release date: | 2001-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Klebsiella Aerogenes Uree, a Nickel-Binding Metallochaperone for Urease Activation.
J.Biol.Chem., 276, 2001
|
|
1TX6
 
 | | trypsin:BBI complex | | Descriptor: | Bowman-Birk type trypsin inhibitor, CALCIUM ION, Trypsin | | Authors: | Song, H.K, Park, E.Y, Kim, J.A, Kim, H.W, Kim, Y.S. | | Deposit date: | 2004-07-02 | | Release date: | 2005-03-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the Bowman-Birk inhibitor from barley seeds in ternary complex with porcine trypsin
J.Mol.Biol., 343, 2004
|
|
1VJS
 
 | |
2DS5
 
 | | Structure of the ZBD in the orthorhomibic crystal from | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit clpX, CALCIUM ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Song, H.K, Park, E.Y, Lee, B.G, Hong, S.B. | | Deposit date: | 2006-06-22 | | Release date: | 2007-02-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis of SspB-tail Recognition by the Zinc Binding Domain of ClpX.
J.Mol.Biol., 367, 2007
|
|
1QZY
 
 | | Human Methionine Aminopeptidase in complex with bengamide inhibitor LAF153 and cobalt | | Descriptor: | (E)-(2R,3R,4S,5R)-3,4,5-TRIHYDROXY-2-METHOXY-8,8-DIMETHYL-NON-6-ENOIC ACID ((3S,6R)-6-HYDROXY-2-OXO-AZEPAN-3-YL)-AMIDE, COBALT (II) ION, Methionine aminopeptidase 2, ... | | Authors: | Eck, M.J, Song, H.K, Morollo, A. | | Deposit date: | 2003-09-18 | | Release date: | 2003-11-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Proteomics-based target identification: bengamides as a new class of methionine aminopeptidase inhibitors.
J.Biol.Chem., 278, 2003
|
|
3RUJ
 
 | |
