8PDZ
 
 | | Recombinant Ena3A L-Type endospore appendages | | Descriptor: | DUF3992 domain-containing protein | | Authors: | Sleutel, M, Remaut, H. | | Deposit date: | 2023-06-13 | | Release date: | 2023-11-08 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | A novel class of ultra-stable endospore appendages decorated with collagen-like tip fibrillae
Biorxiv, 2023
|
|
9H8B
 
 | | Ex vivo cannulae fiber structure of Pyrodictium abyssi | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CanX | | Authors: | Sleutel, M, Remaut, H. | | Deposit date: | 2024-10-28 | | Release date: | 2024-12-18 | | Last modified: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Donor Strand Complementation and Calcium Ion Coordination Drive the Chaperone-free Polymerization of Archaeal Cannulae.
Biorxiv, 2024
|
|
9HZE
 
 | |
9I65
 
 | | Recombinant F-ENA-2 fibers | | Descriptor: | DUF4183 domain-containing protein | | Authors: | Sleutel, M, Remaut, H. | | Deposit date: | 2025-01-29 | | Release date: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM analysis of the Bacillus thuringiensis extrasporal matrix identifies F-ENA as a widespread family of endospore appendages across the Firmicutes phylum.
Biorxiv, 2025
|
|
9I0Y
 
 | | Recombinant Ena2A fibers | | Descriptor: | DUF3992 domain-containing protein | | Authors: | Sleutel, M, Remaut, H. | | Deposit date: | 2025-01-15 | | Release date: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Cryo-EM analysis of the Bacillus thuringiensis extrasporal matrix identifies F-ENA as a widespread family of endospore appendages across the Firmicutes phylum.
Biorxiv, 2025
|
|
7BJZ
 
 | | GLUCOSE ISOMERASE S171W in H32 | | Descriptor: | MANGANESE (II) ION, Xylose isomerase | | Authors: | Sleutel, M. | | Deposit date: | 2021-01-14 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Nucleation of protein mesocrystals via oriented attachment.
Nat Commun, 12, 2021
|
|
8C50
 
 | |
9H38
 
 | |
9H3D
 
 | |
7A02
 
 | | Bacillus endospore appendages form a novel family of disulfide-linked pili | | Descriptor: | DUF3992 domain-containing protein | | Authors: | Pradhan, B, Liedtke, J, Sleutel, M, Lindback, T, Llarena, A.K, Brynildsrud, O, Aspholm, M, Remaut, H. | | Deposit date: | 2020-08-06 | | Release date: | 2021-04-28 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Endospore Appendages: a novel pilus superfamily from the endospores of pathogenic Bacilli.
Embo J., 40, 2021
|
|
4ZV6
 
 | | Crystal structure of the artificial alpharep-7 octarellinV.1 complex | | Descriptor: | AlphaRep-7, Octarellin V.1 | | Authors: | Figueroa, M, Sleutel, M, Urvoas, A, Valerio-Lepiniec, M, Minard, P, Martial, J.A, van de Weerdt, C. | | Deposit date: | 2015-05-18 | | Release date: | 2016-05-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | The unexpected structure of the designed protein Octarellin V.1 forms a challenge for protein structure prediction tools.
J.Struct.Biol., 195, 2016
|
|
5BOP
 
 | | Crystal structure of the artificial nanobody octarellinV.1 complex | | Descriptor: | Nanobody, Octarellin V.1 | | Authors: | Figueroa, M, Sleutel, M, Pardon, E, Steyaert, J, Martial, J.A, van de Weerdt, C. | | Deposit date: | 2015-05-27 | | Release date: | 2016-05-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The unexpected structure of the designed protein Octarellin V.1 forms a challenge for protein structure prediction tools.
J.Struct.Biol., 195, 2016
|
|
4A8I
 
 | | Protein crystallization and microgravity: glucose isomerase crystals grown during the PCDF-PROTEIN mission | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, XYLOSE ISOMERASE | | Authors: | Decanniere, K, Patino-Lopez, L.-D, Sleutel, M, Evrard, C, Van De Weerdt, C, Haumont, E, Gavira, J.A, Otalora, F, Maes, D. | | Deposit date: | 2011-11-21 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Protein Crystallization and Microgravity: Glucose Isomerase Crystals Grown During the Pcdf-Protein Mission
To be Published
|
|
4A8N
 
 | | Protein crystallization and microgravity: glucose isomerase crystals grown during the PCDF-PROTEIN mission | | Descriptor: | COBALT (II) ION, GLYCEROL, XYLOSE ISOMERASE | | Authors: | Decanniere, K, Patino-Lopez, L.-D, Sleutel, M, Evrard, C, Van De Weerdt, C, Haumont, E, Gavira, J.A, Otalora, F, Maes, D. | | Deposit date: | 2011-11-21 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Protein Crystallization and Microgravity: Glucose Isomerase Crystals Grown During the Pcdf-Protein Mission
To be Published
|
|
4A8R
 
 | | Protein crystallization and microgravity: glucose isomerase crystals grown during the PCDF-PROTEIN mission | | Descriptor: | COBALT (II) ION, GLYCEROL, XYLOSE ISOMERASE | | Authors: | Decanniere, K, Patino-Lopez, L.-D, Sleutel, M, Evrard, C, Van De Weerdt, C, Haumont, E, Gavira, J.A, Otalora, F, Maes, D. | | Deposit date: | 2011-11-21 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Protein Crystallization and Microgravity: Glucose Isomerase Crystals Grown During the Pcdf-Protein Mission
To be Published
|
|
4A8L
 
 | | Protein crystallization and microgravity: glucose isomerase crystals grown during the PCDF-PROTEIN mission | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, XYLOSE ISOMERASE | | Authors: | Decanniere, K, Patino-Lopez, L.-D, Sleutel, M, Evrard, C, Van De Weerdt, C, Haumont, E, Gavira, J.A, Otalora, F, Maes, D. | | Deposit date: | 2011-11-21 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Protein Crystallization and Microgravity: Glucose Isomerase Crystals Grown During the Pcdf-Protein Mission
To be Published
|
|
9GK2
 
 | |
2MF2
 
 | |
4MZM
 
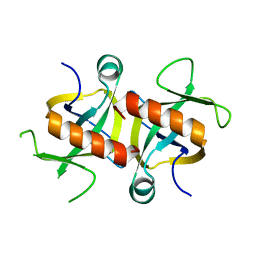 | | MazF from S. aureus crystal form I, P212121, 2.1 A | | Descriptor: | mRNA interferase MazF | | Authors: | Zorzini, V, Loris, R, van Nuland, N.A.J, Cheung, A. | | Deposit date: | 2013-09-30 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and biophysical characterization of Staphylococcus aureus SaMazF shows conservation of functional dynamics.
Nucleic Acids Res., 42, 2014
|
|
4MZP
 
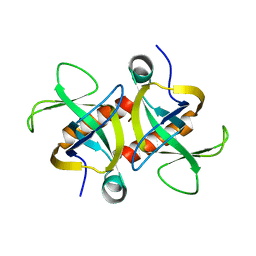 | | MazF from S. aureus crystal form III, C2221, 2.7 A | | Descriptor: | MazF mRNA interferase | | Authors: | Zorzini, V, Loris, R, van Nuland, N.A.J, Cheung, A. | | Deposit date: | 2013-09-30 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Structural and biophysical characterization of Staphylococcus aureus SaMazF shows conservation of functional dynamics.
Nucleic Acids Res., 42, 2014
|
|
4MZT
 
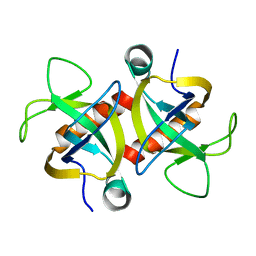 | | MazF from S. aureus crystal form II, C2221, 2.3 A | | Descriptor: | MazF mRNA interferase | | Authors: | Zorzini, V, Loris, R, van Nuland, N.A.J, Cheung, A. | | Deposit date: | 2013-09-30 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Structural and biophysical characterization of Staphylococcus aureus SaMazF shows conservation of functional dynamics.
Nucleic Acids Res., 42, 2014
|
|
4LHN
 
 | |
4LHL
 
 | |
4LHK
 
 | |
