1A0A
 
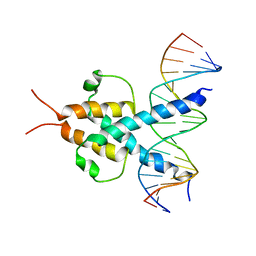 | | PHOSPHATE SYSTEM POSITIVE REGULATORY PROTEIN PHO4/DNA COMPLEX | | Descriptor: | DNA (5'-D(*CP*TP*AP*GP*TP*CP*CP*CP*AP*CP*GP*TP*GP*TP*GP*AP*G )-3'), DNA (5'-D(*CP*TP*CP*AP*CP*AP*CP*GP*TP*GP*GP*GP*AP*CP*TP*AP*G )-3'), PROTEIN (PHOSPHATE SYSTEM POSITIVE REGULATORY PROTEIN PHO4) | | Authors: | Shimizu, T, Toumoto, A, Ihara, K, Shimizu, M, Kyogoku, Y, Ogawa, N, Oshima, Y, Hakoshima, T. | | Deposit date: | 1997-11-27 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of PHO4 bHLH domain-DNA complex: flanking base recognition.
EMBO J., 16, 1997
|
|
5EIO
 
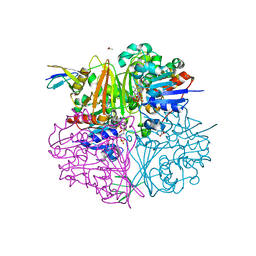 | | Crystal structure of LysY from Thermus thermophilus complexed with NADP+ and LysW-gamma-aminoadipic semialdehyde | | Descriptor: | ACETIC ACID, N-acetyl-gamma-glutamyl-phosphate/N-acetyl-gamma-aminoadipyl-phosphate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Shimizu, T, Tomita, T, Nishiyama, M. | | Deposit date: | 2015-10-30 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the LysYLysW Complex from Thermus thermophilus.
J.Biol.Chem., 291, 2016
|
|
5EIN
 
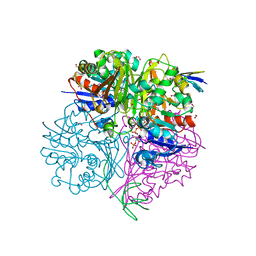 | | Crystal structure of C148A mutant of LysY from Thermus thermophilus in complex with NADP+ and LysW-gamma-aminoadipic acid | | Descriptor: | Alpha-aminoadipate carrier protein LysW, FORMIC ACID, N-acetyl-gamma-glutamyl-phosphate/N-acetyl-gamma-aminoadipyl-phosphate reductase, ... | | Authors: | Shimizu, T, Tomita, T, Nishiyama, M. | | Deposit date: | 2015-10-30 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the LysYLysW Complex from Thermus thermophilus.
J.Biol.Chem., 291, 2016
|
|
1KCD
 
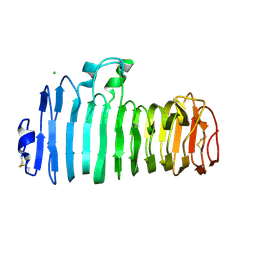 | | Endopolygalacturonase I from Stereum purpureum complexed with two galacturonate at 1.15 A resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ENDOPOLYGALACTURONASE, ... | | Authors: | Shimizu, T, Nakatsu, T, Miyairi, K, Okuno, T, Kato, H. | | Deposit date: | 2001-11-08 | | Release date: | 2002-06-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Active-site architecture of endopolygalacturonase I from Stereum purpureum revealed by crystal structures in native and ligand-bound forms at atomic resolution.
Biochemistry, 41, 2002
|
|
1KCC
 
 | | Endopolygalacturonase I from Stereum purpureum complexed with a galacturonate at 1.00 A resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ENDOPOLYGALACTURONASE, ... | | Authors: | Shimizu, T, Nakatsu, T, Miyairi, K, Okuno, T, Kato, H. | | Deposit date: | 2001-11-08 | | Release date: | 2002-06-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Active-site architecture of endopolygalacturonase I from Stereum purpureum revealed by crystal structures in native and ligand-bound forms at atomic resolution.
Biochemistry, 41, 2002
|
|
1K5C
 
 | | Endopolygalacturonase I from Stereum purpureum at 0.96 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ENDOPOLYGALACTURONASE, ... | | Authors: | Shimizu, T, Nakatsu, T, Miyairi, K, Okuno, T, Kato, H. | | Deposit date: | 2001-10-10 | | Release date: | 2002-06-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Active-site architecture of endopolygalacturonase I from Stereum purpureum revealed by crystal structures in native and ligand-bound forms at atomic resolution.
Biochemistry, 41, 2002
|
|
5HN4
 
 | | Crystal structure of beta-decarboxylating dehydrogenase (TK0280) from Thermococcus kodakarensis complexed with Mn and homoisocitrate | | Descriptor: | (1R,2S)-1-hydroxybutane-1,2,4-tricarboxylic acid, Homoisocitrate dehydrogenase, IMIDAZOLE, ... | | Authors: | Shimizu, T, Tomita, T, Nishiyama, M. | | Deposit date: | 2016-01-18 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structure and function of an ancestral-type beta-decarboxylating dehydrogenase from Thermococcus kodakarensis
Biochem. J., 474, 2017
|
|
5HN6
 
 | | Crystal structure of beta-decarboxylating dehydrogenase (TK0280) from Thermococcus kodakarensis complexed with Mn and 3-isopropylmalate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-ISOPROPYLMALIC ACID, Homoisocitrate dehydrogenase, ... | | Authors: | Shimizu, T, Tomita, T, Nishiyama, M. | | Deposit date: | 2016-01-18 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and function of an ancestral-type beta-decarboxylating dehydrogenase from Thermococcus kodakarensis
Biochem. J., 474, 2017
|
|
5HN5
 
 | | Crystal structure of beta-decarboxylating dehydrogenase (TK0280) from Thermococcus kodakarensis complexed with Mn and isocitrate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Homoisocitrate dehydrogenase, ISOCITRIC ACID, ... | | Authors: | Shimizu, T, Tomita, T, Nishiyama, M. | | Deposit date: | 2016-01-18 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure and function of an ancestral-type beta-decarboxylating dehydrogenase from Thermococcus kodakarensis
Biochem. J., 474, 2017
|
|
5HN3
 
 | |
1ISN
 
 | | Crystal structure of merlin FERM domain | | Descriptor: | merlin | | Authors: | Shimizu, T, Seto, A, Maita, N, Hamada, K, Tsukita, S, Tsukita, S, Hakoshima, T. | | Deposit date: | 2001-12-13 | | Release date: | 2002-04-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for neurofibromatosis type 2. Crystal structure of the merlin FERM domain.
J.Biol.Chem., 277, 2002
|
|
1DPF
 
 | | CRYSTAL STRUCTURE OF A MG-FREE FORM OF RHOA COMPLEXED WITH GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, RHOA | | Authors: | Shimizu, T, Ihara, K, Maesaki, R, Kuroda, S, Kaibuchi, K, Hakoshima, T. | | Deposit date: | 1999-12-27 | | Release date: | 2000-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An open conformation of switch I revealed by the crystal structure of a Mg2+-free form of RHOA complexed with GDP. Implications for the GDP/GTP exchange mechanism.
J.Biol.Chem., 275, 2000
|
|
1VMO
 
 | | CRYSTAL STRUCTURE OF VITELLINE MEMBRANE OUTER LAYER PROTEIN I (VMO-I): A FOLDING MOTIF WITH HOMOLOGOUS GREEK KEY STRUCTURES RELATED BY AN INTERNAL THREE-FOLD SYMMETRY | | Descriptor: | VITELLINE MEMBRANE OUTER LAYER PROTEIN I | | Authors: | Shimizu, T, Vassylyev, D.G, Kido, S, Doi, Y, Morikawa, K. | | Deposit date: | 1994-01-06 | | Release date: | 1994-05-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of vitelline membrane outer layer protein I (VMO-I): a folding motif with homologous Greek key structures related by an internal three-fold symmetry.
EMBO J., 13, 1994
|
|
1UIX
 
 | | Coiled-coil structure of the RhoA-binding domain in Rho-kinase | | Descriptor: | Rho-associated kinase | | Authors: | Shimizu, T, Ihara, K, Maesaki, R, Amano, M, Kaibuchi, K, Hakoshima, T. | | Deposit date: | 2003-07-23 | | Release date: | 2003-10-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Parallel coiled-coil association of the RhoA-binding domain in Rho-kinase
J.Biol.Chem., 278, 2003
|
|
7YTX
 
 | | Crystal structure of TLR8 in complex with its antagonist | | Descriptor: | (2R,6R)-4-(8-cyanoquinolin-5-yl)-N-[(3S,4R)-4-fluoranylpyrrolidin-3-yl]-6-methyl-morpholine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shimizu, T, Sakaniwa, K. | | Deposit date: | 2022-08-16 | | Release date: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A novel Toll-like receptor 7/8-specific antagonist E6742 ameliorates clinically relevant disease parameters in murine models of lupus.
Eur.J.Pharmacol., 957, 2023
|
|
5GIC
 
 | | Crystal structure of VDR in complex with DLAM-2P | | Descriptor: | (3~{R},5~{S})-5-[(2~{R})-2-[(1~{R},3~{a}~{S},4~{Z},7~{a}~{R})-7~{a}-methyl-4-[(2~{E})-2-[(3~{S},5~{R})-2-methylidene-3,5-bis(oxidanyl)cyclohexylidene]ethylidene]-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-1-yl]propyl]-3-methyl-3-oxidanyl-1-(2-phenylethyl)pyrrolidin-2-one, SRC1, Vitamin D3 receptor | | Authors: | Shimizu, T, Asano, L. | | Deposit date: | 2016-06-23 | | Release date: | 2016-12-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.352 Å) | | Cite: | Regulation of the vitamin D receptor by vitamin D lactam derivatives.
Febs Lett., 590, 2016
|
|
3W0I
 
 | | Crystal Structure of Rat VDR Ligand Binding Domain in Complex with Novel Nonsecosteroidal Ligands | | Descriptor: | (2S)-3-{4-[3-(4-{[(2R)-2-hydroxy-3,3-dimethylbutyl]oxy}phenyl)pentan-3-yl]phenoxy}propane-1,2-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 Receptor | | Authors: | Shimizu, T, Asano, L, Kuwabara, N, Ito, I, Waku, T, Yanagisawa, J, Miyachi, H. | | Deposit date: | 2012-10-30 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for vitamin D receptor agonism by novel non-secosteroidal ligands.
Febs Lett., 587, 2013
|
|
3W0H
 
 | | Crystal Structure of Rat VDR Ligand Binding Domain in Complex with Novel Nonsecosteroidal Ligands | | Descriptor: | (2S)-3-{4-[4-(4-{[(2R)-2-hydroxy-3,3-dimethylbutyl]oxy}phenyl)heptan-4-yl]phenoxy}propane-1,2-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Shimizu, T, Asano, L, Kuwabara, N, Ito, I, Waku, T, Yanagisawa, J, Miyachi, H. | | Deposit date: | 2012-10-30 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for vitamin D receptor agonism by novel non-secosteroidal ligands.
Febs Lett., 587, 2013
|
|
3W0G
 
 | | Crystal Structure of Rat VDR Ligand Binding Domain in Complex with Novel Nonsecosteroidal Ligands | | Descriptor: | (2S)-3-{4-[2-(4-{[(2R)-2-hydroxy-3,3-dimethylbutyl]oxy}phenyl)propan-2-yl]phenoxy}propane-1,2-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Shimizu, T, Asano, L, Kuwabara, N, Ito, I, Waku, T, Yanagisawa, J, Miyachi, H. | | Deposit date: | 2012-10-30 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural basis for vitamin D receptor agonism by novel non-secosteroidal ligands.
Febs Lett., 587, 2013
|
|
3W0J
 
 | | Crystal Structure of Rat VDR Ligand Binding Domain in Complex with Novel Nonsecosteroidal Ligands | | Descriptor: | (2S)-3-{4-[2-(4-{[(2R)-2-hydroxy-3,3-dimethylbutyl]oxy}-3-methylphenyl)propan-2-yl]-2-methylphenoxy}propane-1,2-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 Receptor | | Authors: | Shimizu, T, Asano, L, Kuwabara, N, Ito, I, Waku, T, Yanagisawa, J, Miyachi, H. | | Deposit date: | 2012-10-30 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural basis for vitamin D receptor agonism by novel non-secosteroidal ligands.
Febs Lett., 587, 2013
|
|
3B0T
 
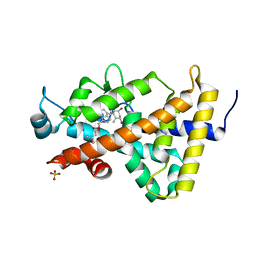 | | Human VDR ligand binding domain in complex with maxacalcitol | | Descriptor: | (1S,3R,5Z,7E,14beta,17alpha,20S)-20-(3-hydroxy-3-methylbutoxy)-9,10-secopregna-5,7,10-triene-1,3-diol, SULFATE ION, Vitamin D3 receptor | | Authors: | Hishiki, A, Hashimoto, H, Sato, M, Shimizu, T. | | Deposit date: | 2011-06-14 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Human VDR ligand binding domain in complex with maxacalcitol
To be Published
|
|
3B0R
 
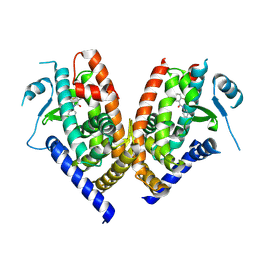 | |
2A0B
 
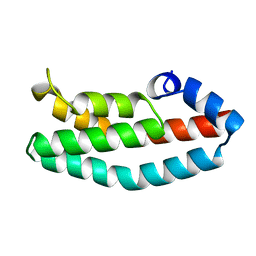 | | HISTIDINE-CONTAINING PHOSPHOTRANSFER DOMAIN OF ARCB FROM ESCHERICHIA COLI | | Descriptor: | HPT DOMAIN, ZINC ION | | Authors: | Kato, M, Mizuno, T, Shimizu, T, Hakoshima, T. | | Deposit date: | 1998-04-02 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Refined structure of the histidine-containing phosphotransfer (HPt) domain of the anaerobic sensor kinase ArcB from Escherichia coli at 1.57 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
5DST
 
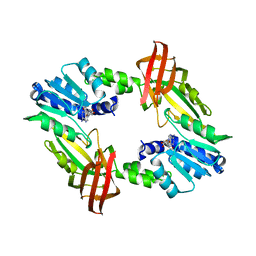 | |
1A0B
 
 | | HISTIDINE-CONTAINING PHOSPHOTRANSFER DOMAIN OF ARCB FROM ESCHERICHIA COLI | | Descriptor: | AEROBIC RESPIRATION CONTROL SENSOR PROTEIN ARCB, ZINC ION | | Authors: | Kato, M, Mizuno, T, Shimizu, T, Hakoshima, T. | | Deposit date: | 1997-11-27 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Insights into multistep phosphorelay from the crystal structure of the C-terminal HPt domain of ArcB.
Cell(Cambridge,Mass.), 88, 1997
|
|
