1C28
 
 | |
1NCI
 
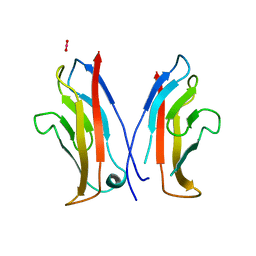 | | STRUCTURAL BASIS OF CELL-CELL ADHESION BY CADHERINS | | Descriptor: | N-CADHERIN, URANYL (VI) ION | | Authors: | Shapiro, L, Fannon, A.M, Kwong, P.D, Thompson, A, Lehmann, M.S, Grubel, G, Legrand, J.-F, Als-Nielsen, J, Colman, D.R, Hendrickson, W.A. | | Deposit date: | 1995-03-23 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of cell-cell adhesion by cadherins.
Nature, 374, 1995
|
|
1NEU
 
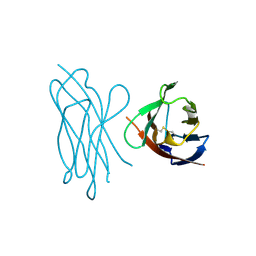 | | STRUCTURE OF MYELIN MEMBRANE ADHESION MOLECULE P0 | | Descriptor: | MYELIN P0 PROTEIN | | Authors: | Shapiro, L, Doyle, J.P, Hensley, P, Colman, D.R, Hendrickson, W.A. | | Deposit date: | 1996-09-24 | | Release date: | 1997-05-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the extracellular domain from P0, the major structural protein of peripheral nerve myelin.
Neuron, 17, 1996
|
|
1NCH
 
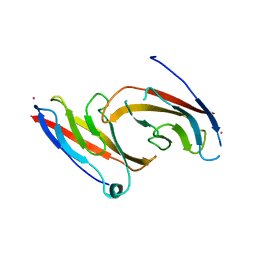 | | STRUCTURAL BASIS OF CELL-CELL ADHESION BY CADHERINS | | Descriptor: | N-CADHERIN, YTTERBIUM (III) ION | | Authors: | Shapiro, L, Fannon, A.M, Kwong, P.D, Thompson, A, Lehmann, M.S, Grubel, G, Legrand, J.-F, Als-Nielsen, J, Colman, D.R, Hendrickson, W.A. | | Deposit date: | 1995-03-23 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of cell-cell adhesion by cadherins.
Nature, 374, 1995
|
|
1NCG
 
 | | STRUCTURAL BASIS OF CELL-CELL ADHESION BY CADHERINS | | Descriptor: | N-CADHERIN, YTTERBIUM (III) ION | | Authors: | Shapiro, L, Fannon, A.M, Kwong, P.D, Thompson, A, Lehmann, M.S, Grubel, G, Legrand, J.-F, Als-Nielsen, J, Colman, D.R, Hendrickson, W.A. | | Deposit date: | 1995-03-23 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of cell-cell adhesion by cadherins.
Nature, 374, 1995
|
|
3K6F
 
 | |
3K5S
 
 | |
1C3H
 
 | | ACRP30 CALCIUM COMPLEX | | Descriptor: | 30 KD ADIPOCYTE COMPLEMENT-RELATED PROTEIN PRECURSOR, CALCIUM ION | | Authors: | Shapiro, L, Boggon, T, Scherer, P. | | Deposit date: | 1999-07-27 | | Release date: | 2003-12-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | ACRP30 calcium complex
To Be Published
|
|
6WUD
 
 | |
6WU7
 
 | |
3K6I
 
 | |
3K5R
 
 | |
3K6D
 
 | |
3BL8
 
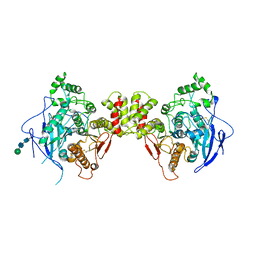 | | Crystal structure of the extracellular domain of neuroligin 2A from mouse | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Neuroligin-2, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jin, X, Koehnke, J, Shapiro, L. | | Deposit date: | 2007-12-10 | | Release date: | 2008-02-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the extracellular cholinesterase-like domain from neuroligin-2.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
7N5H
 
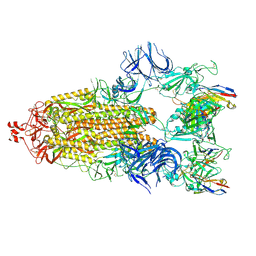 | | Cryo-EM structure of broadly neutralizing antibody 2-36 in complex with prefusion SARS-CoV-2 spike glycoprotein | | Descriptor: | 2-36 Fab heavy chain, 2-36 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Casner, R.G, Cerutti, G, Shapiro, L. | | Deposit date: | 2021-06-05 | | Release date: | 2021-11-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | A monoclonal antibody that neutralizes SARS-CoV-2 variants, SARS-CoV, and other sarbecoviruses.
Emerg Microbes Infect, 11, 2022
|
|
9EKF
 
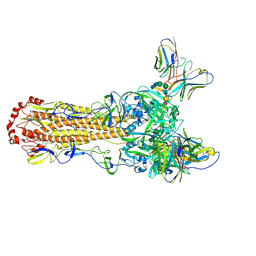 | | CryoEM structure of H5N1 A/Texas/37/2024 HA bound to Fab 65C6 and an auto glycan occupying the receptor-binding site | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 65C6 Heavy Chain, Fab 65C6 Light Chain, ... | | Authors: | Morano, N.C, Shapiro, L, Kwong, P.D. | | Deposit date: | 2024-12-02 | | Release date: | 2024-12-18 | | Last modified: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structure of a zoonotic H5N1 hemagglutinin reveals a receptor-binding site occupied by an auto-glycan.
Structure, 33, 2025
|
|
2QRD
 
 | | Crystal Structure of the Adenylate Sensor from AMP-activated Protein Kinase in complex with ADP and ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Protein C1556.08c, ... | | Authors: | Jin, X, Townley, R, Shapiro, L. | | Deposit date: | 2007-07-28 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural Insight into AMPK Regulation: ADP Comes into Play.
Structure, 15, 2007
|
|
2QRE
 
 | | Crystal structure of the adenylate sensor from AMP-activated protein kinase in complex with 5-aminoimidazole-4-carboxamide 1-beta-D-ribofuranotide (ZMP) | | Descriptor: | AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, Protein C1556.08c, SNF1-like protein kinase ssp2, ... | | Authors: | Jin, X, Townley, R, Shapiro, L. | | Deposit date: | 2007-07-28 | | Release date: | 2007-10-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural Insight into AMPK Regulation: ADP Comes into Play.
Structure, 15, 2007
|
|
2QR1
 
 | | Crystal structure of the adenylate sensor from AMP-activated protein kinase in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Protein C1556.08c, SNF1-like protein kinase ssp2, ... | | Authors: | Jin, X, Townley, R, Shapiro, L. | | Deposit date: | 2007-07-27 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insight into AMPK Regulation: ADP Comes into Play.
Structure, 15, 2007
|
|
1I7E
 
 | | C-Terminal Domain Of Mouse Brain Tubby Protein bound to Phosphatidylinositol 4,5-bis-phosphate | | Descriptor: | L-ALPHA-GLYCEROPHOSPHO-D-MYO-INOSITOL-4,5-BIS-PHOSPHATE, TUBBY PROTEIN | | Authors: | Santagata, S, Boggon, T.J, Baird, C.L, Shan, W.S, Shapiro, L. | | Deposit date: | 2001-03-08 | | Release date: | 2001-06-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | G-protein signaling through tubby proteins.
Science, 292, 2001
|
|
9DD7
 
 | | Cryo-EM structure of neutralizing human antibody D48 in complex with HSV-1 glycoprotein B trimer gB-Ecto.516P.531E.DS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, D48 heavy chain, ... | | Authors: | Roark, R.S, Shapiro, L, Kwong, P.D. | | Deposit date: | 2024-08-27 | | Release date: | 2025-09-10 | | Last modified: | 2025-11-12 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Prefusion structure, evasion and neutralization of HSV-1 glycoprotein B.
Nat Microbiol, 10, 2025
|
|
9DDA
 
 | | Cryo-EM structure of gB-Ecto.516P.531E.DS, a prefusion-stabilized HSV-1 glycoprotein B extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein B, ... | | Authors: | Roark, R.S, Shapiro, L, Kwong, P.D. | | Deposit date: | 2024-08-27 | | Release date: | 2025-09-10 | | Last modified: | 2025-11-12 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Prefusion structure, evasion and neutralization of HSV-1 glycoprotein B.
Nat Microbiol, 10, 2025
|
|
9DD8
 
 | | Cryo-EM structure of neutralizing human antibody D48 in complex with HSV-1 glycoprotein B trimer gB-Ecto.516P | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, D48 heavy chain, ... | | Authors: | Roark, R.S, Shapiro, L, Kwong, P.D. | | Deposit date: | 2024-08-27 | | Release date: | 2025-09-10 | | Last modified: | 2025-11-12 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Prefusion structure, evasion and neutralization of HSV-1 glycoprotein B.
Nat Microbiol, 10, 2025
|
|
6VFW
 
 | | Crystal structure of human delta protocadherin 10 EC1-EC4 | | Descriptor: | CALCIUM ION, Protocadherin-10, alpha-D-mannopyranose, ... | | Authors: | Harrison, O.J, Brasch, J, Shapiro, L. | | Deposit date: | 2020-01-06 | | Release date: | 2020-03-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Family-wide Structural and Biophysical Analysis of Binding Interactions among Non-clustered delta-Protocadherins.
Cell Rep, 30, 2020
|
|
6VFQ
 
 | | Crystal structure of monomeric human protocadherin 10 EC1-EC4 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Harrison, O.J, Brasch, J, Shapiro, L. | | Deposit date: | 2020-01-06 | | Release date: | 2020-03-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Family-wide Structural and Biophysical Analysis of Binding Interactions among Non-clustered delta-Protocadherins.
Cell Rep, 30, 2020
|
|
