8UR4
 
 | |
4LGO
 
 | |
4W91
 
 | |
3S82
 
 | |
6VEL
 
 | |
4TVI
 
 | |
6UWW
 
 | |
9NV5
 
 | | Crystal structure of phosphoglycerate mutase from Trichomonas vaginalis | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, GLYCEROL, Phosphoglycerate mutase, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2025-03-20 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of phosphoglycerate mutase from Trichomonas vaginalis
To be published
|
|
9OBA
 
 | |
9MJQ
 
 | |
9OBC
 
 | |
9O68
 
 | |
9O66
 
 | |
9O9R
 
 | |
9OB9
 
 | |
9NSR
 
 | |
9O8N
 
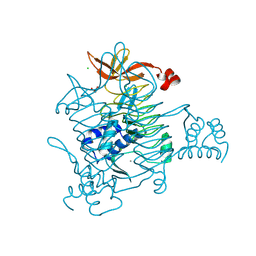 | | Crystal structure of 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase (DapD) from Bordetella pertussis | | Descriptor: | 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2025-04-16 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystal structure of 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase (dapD) from Bordetella pertussis
To be published
|
|
9O57
 
 | |
9O8Y
 
 | | Crystal structure of 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase (DapD) from Bordetella pertussis (succinyl-CoA bound) | | Descriptor: | 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase, CHLORIDE ION, SUCCINYL-COENZYME A | | Authors: | Seattle Structural Genomics Center for Infectious Disease, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2025-04-17 | | Release date: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase (dapD) from Bordetella pertussis (succinyl-CoA bound)
To be published
|
|
9O8J
 
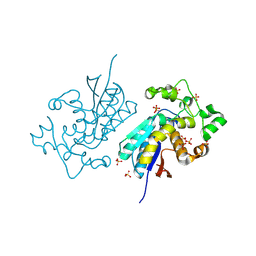 | |
9O90
 
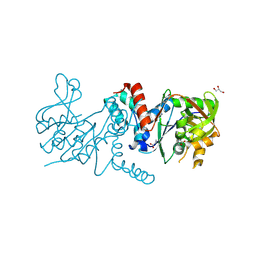 | |
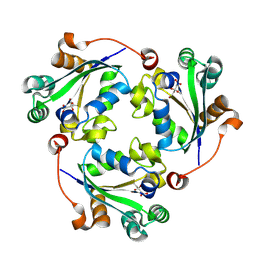
9OD6
 
 | |
9OAN
 
 | |
9OAI
 
 | |
9OAK
 
 | |
