1JXQ
 
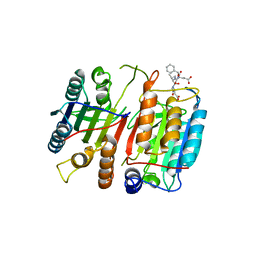 | | Structure of cleaved, CARD domain deleted Caspase-9 | | Descriptor: | Caspase-9, benzoxycarbonyl-Val-Ala-Asp-fluoromethyl ketone Inhibitor | | Authors: | Renatus, M, Stennicke, H.R, Scott, F.L, Liddington, R.C, Salvesen, G.S. | | Deposit date: | 2001-09-08 | | Release date: | 2001-12-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dimer formation drives the activation of the cell death protease caspase 9.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
3ITN
 
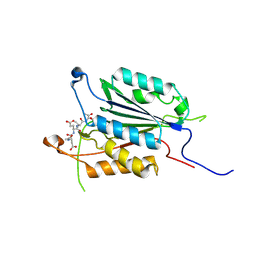 | | Crystal structure of Pseudo-activated Procaspase-3 | | Descriptor: | ACETYL-ASP-GLU-VAL-ASP-CHLOROMETHYL KETONE inhibitor, Caspase-3 | | Authors: | Walters, J, Pop, C, Scott, F.L, Drag, M, Swartz, P.D, Mattos, C, Salvesen, G.S, Clark, A.C. | | Deposit date: | 2009-08-28 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | A constitutively active and uninhibitable caspase-3 zymogen efficiently induces apoptosis.
Biochem.J., 424, 2009
|
|
3V2W
 
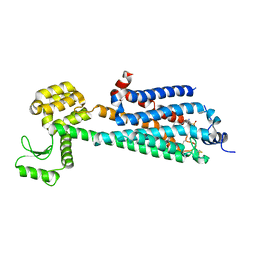 | | Crystal Structure of a Lipid G protein-Coupled Receptor at 3.35A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Sphingosine 1-phosphate receptor 1, Lysozyme chimera, ... | | Authors: | Hanson, M.A, Roth, C.B, Jo, E, Griffith, M.T, Scott, F.L, Reinhart, G, Desale, H, Clemons, B, Cahalan, S.M, Schuerer, S.C, Sanna, M.G, Han, G.W, Kuhn, P, Rosen, H, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2011-12-12 | | Release date: | 2012-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Crystal structure of a lipid G protein-coupled receptor.
Science, 335, 2012
|
|
3V2Y
 
 | | Crystal Structure of a Lipid G protein-Coupled Receptor at 2.80A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Sphingosine 1-phosphate receptor 1, Lysozyme chimera (E.C.3.2.1.17), ... | | Authors: | Hanson, M.A, Roth, C.B, Jo, E, Griffith, M.T, Scott, F.L, Reinhart, G, Desale, H, Clemons, B, Cahalan, S.M, Schuerer, S.C, Sanna, M.G, Han, G.W, Kuhn, P, Rosen, H, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2011-12-12 | | Release date: | 2012-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a lipid G protein-coupled receptor.
Science, 335, 2012
|
|
3EZQ
 
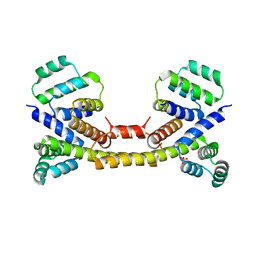 | | Crystal Structure of the Fas/FADD Death Domain Complex | | Descriptor: | Protein FADD, SODIUM ION, SULFATE ION, ... | | Authors: | Schwarzenbacher, R, Robinson, H, Stec, B, Riedl, S.J. | | Deposit date: | 2008-10-23 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | The Fas-FADD death domain complex structure unravels signalling by receptor clustering
Nature, 457, 2009
|
|
