1S4S
 
 | | Reaction Intermediate in the Photocycle of PYP, intermediate occupied between 100 micro-seconds to 5 milli-seconds | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Schmidt, M, Pahl, R, Srajer, V, Anderson, S, Ren, Z, Ihee, H, Rajagopal, S, Moffat, K. | | Deposit date: | 2004-01-17 | | Release date: | 2004-04-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein kinetics: Structures of intermediates and reaction mechanism from time-resolved x-ray data
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
5AEF
 
 | | Electron cryo-microscopy of an Abeta(1-42)amyloid fibril | | Descriptor: | AMYLOID BETA A4 PROTEIN | | Authors: | Schmidt, M, Rohou, A, Lasker, K, Yadav, J.K, Schiene-Fischer, C, Fandrich, M, Grigorieff, N. | | Deposit date: | 2015-08-29 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Peptide Dimer Structure in an Abeta(1-42) Fibril Visualized with Cryo-Em
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1AVM
 
 | |
1BS3
 
 | | P.SHERMANII SOD(FE+3) FLUORIDE | | Descriptor: | FE (III) ION, FLUORIDE ION, SUPEROXIDE DISMUTASE | | Authors: | Schmidt, M. | | Deposit date: | 1998-08-31 | | Release date: | 1999-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Manipulating the coordination mumber of the ferric iron within the cambialistic superoxide dismutase of Propionibacterium shermanii by changing the pH-value A crystallographic analysis
Eur.J.Biochem., 262, 1999
|
|
1BSM
 
 | | P.SHERMANII SOD(FE+3) 140K PH8 | | Descriptor: | FE (III) ION, SUPEROXIDE DISMUTASE | | Authors: | Schmidt, M. | | Deposit date: | 1998-08-28 | | Release date: | 1999-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Manipulating the coordination mumber of the ferric iron within the cambialistic superoxide dismutase of Propionibacterium shermanii by changing the pH-value A crystallographic analysis
Eur.J.Biochem., 262, 1999
|
|
1BT8
 
 | | P.SHERMANII SOD(FE+3) PH 10.0 | | Descriptor: | FE (III) ION, SUPEROXIDE DISMUTASE | | Authors: | Schmidt, M. | | Deposit date: | 1998-09-01 | | Release date: | 1999-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Manipulating the coordination mumber of the ferric iron within the cambialistic superoxide dismutase of Propionibacterium shermanii by changing the pH-value A crystallographic analysis
Eur.J.Biochem., 262, 1999
|
|
2BW9
 
 | | Laue Structure of L29W MbCO | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Schmidt, M, Nienhaus, K, Pahl, R, Krasselt, A, Anderson, S, Parak, F, Nienhaus, G.U, Srajer, V. | | Deposit date: | 2005-07-13 | | Release date: | 2005-07-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Ligand Migration Pathway and Protein Dynamics in Myoglobin: A Time-Resolved Crystallographic Study on L29W Mbco.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2C7K
 
 | | Laue structure of phycoerythrocyanin from Mastigocladus laminosus | | Descriptor: | BILIVERDINE IX ALPHA, PHYCOCYANOBILIN, PHYCOERYTHROCYANIN ALPHA CHAIN, ... | | Authors: | Schmidt, M, Krasselt, A, Reuter, W. | | Deposit date: | 2005-11-24 | | Release date: | 2006-01-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Local Protein Flexibility as a Prerequisite for Reversible Chromophore Isomerization in Alpha-Phycoerythrocyanin
Biochim.Biophys.Acta, 1764, 2006
|
|
2C7J
 
 | | Phycoerythrocyanin from Mastigocladus laminosus, 295 K, 3.0 A | | Descriptor: | BILIVERDINE IX ALPHA, PHYCOCYANOBILIN, PHYCOERYTHROCYANIN ALPHA CHAIN, ... | | Authors: | Schmidt, M, Krasselt, A, Reuter, W. | | Deposit date: | 2005-11-24 | | Release date: | 2006-01-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Local Protein Flexibility as a Prerequisite for Reversible Chromophore Isomerization in Alfa- Phycoerythrocyanin
Biochim.Biophys.Acta, 1764, 2006
|
|
2C7L
 
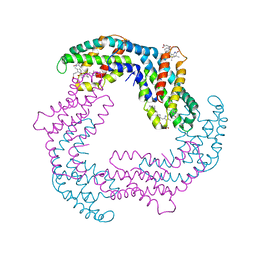 | | Low temperature structure of phycoerythrocyanin from Mastigocladus laminosus | | Descriptor: | BILIVERDINE IX ALPHA, PHYCOCYANOBILIN, PHYCOERYTHROCYANIN ALPHA CHAIN, ... | | Authors: | Schmidt, M, Krasselt, A, Reuter, W. | | Deposit date: | 2005-11-25 | | Release date: | 2006-01-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Local Protein Flexibility as a Prerequisite for Reversible Chromophore Isomerization in Alpha-Phycoerythrocyanin
Biochim.Biophys.Acta, 1764, 2006
|
|
2BWH
 
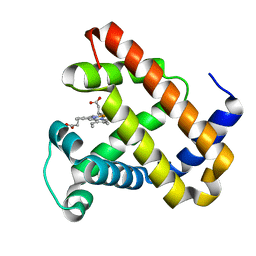 | | Laue Structure of a Short Lived State of L29W Myoglobin | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Schmidt, M, Nienhaus, K, Pahl, R, Krasselt, A, Anderson, S, Parak, F, Nienhaus, G.U, Srajer, V. | | Deposit date: | 2005-07-14 | | Release date: | 2005-07-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ligand migration pathway and protein dynamics in myoglobin: a time-resolved crystallographic study on L29W MbCO.
Proc. Natl. Acad. Sci. U.S.A., 102, 2005
|
|
1S4R
 
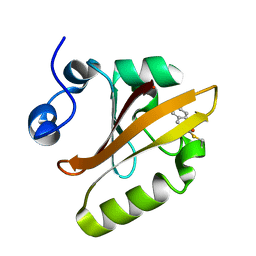 | | Structure of a reaction intermediate in the photocycle of PYP extracted by a SVD-driven analysis | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Schmidt, M, Pahl, R, Srajer, V, Anderson, S, Ren, Z, Ihee, H, Rajagopal, S, Moffat, K. | | Deposit date: | 2004-01-17 | | Release date: | 2004-04-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein kinetics: Structures of intermediates and reaction mechanism from time-resolved x-ray data
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1AR5
 
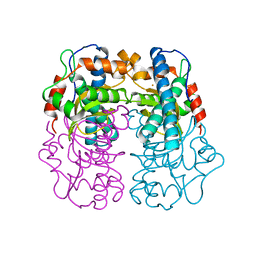 | |
1AR4
 
 | |
7JVZ
 
 | |
2J96
 
 | | The E-configuration of alfa-Phycoerythrocyanin | | Descriptor: | PHYCOERYTHROCYANIN ALPHA CHAIN, PHYCOVIOLOBILIN | | Authors: | Schmidt, M, Patel, A, Zhao, Y, Reuter, W. | | Deposit date: | 2006-11-02 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Basis for the Photochemistry of Alfa-Phycoerythrocyanin
Biochemistry, 46, 2007
|
|
6P73
 
 | | Cytochrome-C-nitrite reductase | | Descriptor: | CALCIUM ION, Cytochrome c-552, HEME C | | Authors: | Schmidt, M, Pacheco, A. | | Deposit date: | 2019-06-04 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Trapping of a Putative Intermediate in the CytochromecNitrite Reductase (ccNiR)-Catalyzed Reduction of Nitrite: Implications for the ccNiR Reaction Mechanism.
J.Am.Chem.Soc., 141, 2019
|
|
8ADE
 
 | |
6BAO
 
 | | Stigmatella aurantiaca phytochrome photosensory core module, wild type | | Descriptor: | 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Photoreceptor-histidine kinase BphP | | Authors: | Schmidt, M, Stojkovic, E. | | Deposit date: | 2017-10-14 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural basis for light control of cell development revealed by crystal structures of a myxobacterial phytochrome.
IUCrJ, 5, 2018
|
|
6BAK
 
 | | The structure of the Stigmatella aurantiaca phytochrome chromophore binding domain T289H mutant | | Descriptor: | 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Photoreceptor-histidine kinase BphP | | Authors: | Schmidt, M, Stojkovic, E. | | Deposit date: | 2017-10-13 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis for light control of cell development revealed by crystal structures of a myxobacterial phytochrome.
IUCrJ, 5, 2018
|
|
7JRI
 
 | |
7JR5
 
 | | Real Time Reaction Intermediates in Stigmatella Bacteriophytochrome P2 | | Descriptor: | 3-[2-[[5-[(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-5-[[(3~{S})-4-ethyl-3-methyl-2-oxidanylidene-1,3-dihydropyrrol-5-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, BENZAMIDINE, Photoreceptor-histidine kinase BphP | | Authors: | Schmidt, M. | | Deposit date: | 2020-08-11 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | High-resolution crystal structures of transient intermediates in the phytochrome photocycle.
Structure, 29, 2021
|
|
6BAF
 
 | | Structure of the chromophore binding domain of Stigmatella aurantiaca phytochrome P1, wild-type | | Descriptor: | 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Photoreceptor-histidine kinase BphP | | Authors: | Schmidt, M, Stojkovic, E. | | Deposit date: | 2017-10-12 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for light control of cell development revealed by crystal structures of a myxobacterial phytochrome.
IUCrJ, 5, 2018
|
|
6BAP
 
 | | Stigmatella aurantiaca bacterial phytochrome PAS-GAF-PHY, T289H mutant | | Descriptor: | 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Photoreceptor-histidine kinase BphP | | Authors: | Schmidt, M, Stojkovic, E. | | Deposit date: | 2017-10-14 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis for light control of cell development revealed by crystal structures of a myxobacterial phytochrome.
IUCrJ, 5, 2018
|
|
6BAY
 
 | | Stigmatella aurantiaca bacterial phytochrome P1, PAS-GAF-PHY T289H mutant, room temperature structure | | Descriptor: | 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Photoreceptor-histidine kinase BphP | | Authors: | Schmidt, M, Stojkovic, E. | | Deposit date: | 2017-10-16 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis for light control of cell development revealed by crystal structures of a myxobacterial phytochrome.
IUCrJ, 5, 2018
|
|
