3JBY
 
 | | Cryo-electron microscopy structure of RAG Paired Complex (C2 symmetry) | | Descriptor: | '-D(P*GP*AP*TP*CP*TP*GP*GP*CP*CP*TP*GP*TP*CP*TP*TP*A)-3', 5'-D(P*CP*AP*CP*AP*GP*TP*GP*CP*TP*AP*CP*AP*GP*AP*C)-3', CALCIUM ION, ... | | Authors: | Ru, H, Chambers, M.G, Fu, T.-M, Tong, A.B, Liao, M, Wu, H. | | Deposit date: | 2015-10-22 | | Release date: | 2015-12-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular Mechanism of V(D)J Recombination from Synaptic RAG1-RAG2 Complex Structures.
Cell(Cambridge,Mass.), 163, 2015
|
|
3JBX
 
 | | Cryo-electron microscopy structure of RAG Signal End Complex (C2 symmetry) | | Descriptor: | 5'-D(*CP*AP*CP*AP*GP*TP*GP*CP*TP*AP*CP*AP*GP*AP*C)-3', 5'-D(*GP*CP*GP*AP*TP*GP*GP*TP*TP*AP*AP*CP*CP*A)-3', 5'-D(P*GP*TP*CP*TP*GP*TP*AP*GP*CP*AP*CP*TP*GP*TP*G)-3', ... | | Authors: | Ru, H, Chambers, M.G, Fu, T.-M, Tong, A.B, Liao, M, Wu, H. | | Deposit date: | 2015-10-22 | | Release date: | 2015-12-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular Mechanism of V(D)J Recombination from Synaptic RAG1-RAG2 Complex Structures.
Cell(Cambridge,Mass.), 163, 2015
|
|
3JBW
 
 | | Cryo-electron microscopy structure of RAG Paired Complex (with NBD, no symmetry) | | Descriptor: | 12-RSS signal end forward strand, 5'-D(P*GP*AP*TP*CP*TP*GP*GP*CP*CP*TP*GP*TP*CP*TP*TP*A)-3', Nicked 12-RSS intermediate reverse strand, ... | | Authors: | Ru, H, Chambers, M.G, Fu, T.-M, Tong, A.B, Liao, M, Wu, H. | | Deposit date: | 2015-10-21 | | Release date: | 2015-12-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Molecular Mechanism of V(D)J Recombination from Synaptic RAG1-RAG2 Complex Structures.
Cell(Cambridge,Mass.), 163, 2015
|
|
3U3Q
 
 | | The S-SAD phased crystal structure of the ecto-domain of Death Receptor 6 (DR6) | | Descriptor: | Tumor necrosis factor receptor superfamily member 21 | | Authors: | Ru, H, Zhao, L.X, Ding, W, Jiao, L.Y, Shaw, N, Zhang, L.G, Hung, L.W, Matsugaki, N, Wakatsuki, S, Liu, Z.J. | | Deposit date: | 2011-10-06 | | Release date: | 2012-05-02 | | Last modified: | 2013-07-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | S-SAD phasing study of death receptor 6 and its solution conformation revealed by SAXS.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3U3S
 
 | | The S-SAD phased crystal structure of the ecto-domain of Death Receptor 6 (DR6) | | Descriptor: | Tumor necrosis factor receptor superfamily member 21 | | Authors: | Ru, H, Zhao, L.X, Ding, W, Jiao, L.Y, Shaw, N, Zhang, L.G, Hung, L.W, Matsugaki, N, Wakatsuki, S, Liu, Z.J. | | Deposit date: | 2011-10-06 | | Release date: | 2012-05-02 | | Last modified: | 2013-07-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | S-SAD phasing study of death receptor 6 and its solution conformation revealed by SAXS.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3U3P
 
 | | The S-SAD phased crystal structure of the ecto-domain of Death Receptor 6 (DR6) | | Descriptor: | Tumor necrosis factor receptor superfamily member 21 | | Authors: | Ru, H, Zhao, L.X, Ding, W, Jiao, L.Y, Shaw, N, Zhang, L.G, Hung, L.W, Matsugaki, N, Wakatsuki, S, Liu, Z.J. | | Deposit date: | 2011-10-06 | | Release date: | 2012-05-02 | | Last modified: | 2013-07-10 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | S-SAD phasing study of death receptor 6 and its solution conformation revealed by SAXS
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3U3V
 
 | | The S-SAD phased crystal structure of the ecto-domain of Death Receptor 6 (DR6) | | Descriptor: | Tumor necrosis factor receptor superfamily member 21 | | Authors: | Ru, H, Zhao, L.X, Ding, W, Jiao, L.Y, Shaw, N, Zhang, L.G, Hung, L.W, Matsugaki, N, Wakatsuki, S, Liu, Z.J. | | Deposit date: | 2011-10-06 | | Release date: | 2012-05-02 | | Last modified: | 2012-07-11 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | S-SAD phasing study of death receptor 6 and its solution conformation revealed by SAXS
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3U3T
 
 | | The S-SAD phased crystal structure of the ecto-domain of Death Receptor 6 (DR6) | | Descriptor: | Tumor necrosis factor receptor superfamily member 21 | | Authors: | Ru, H, Zhao, L.X, Ding, W, Jiao, L.Y, Shaw, N, Zhang, L.G, Hung, L.W, Matsugaki, N, Wakatsuki, S, Liu, Z.J. | | Deposit date: | 2011-10-06 | | Release date: | 2012-05-02 | | Last modified: | 2012-07-11 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | S-SAD phasing study of death receptor 6 and its solution conformation revealed by SAXS
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4JBJ
 
 | | Structural mimicry for functional antagonism | | Descriptor: | Interferon-activable protein 202 | | Authors: | Ru, H, Ni, X, Ma, F, Zhao, L, Ding, W, Hung, L.-W, Shaw, N, Cheng, G, Liu, Z.-J. | | Deposit date: | 2013-02-19 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.692 Å) | | Cite: | Structural basis for termination of AIM2-mediated signaling by p202
Cell Res., 23, 2013
|
|
4JBK
 
 | | Molecular basis for abrogation of activation of pro-inflammatory cytokines | | Descriptor: | DNA (5'-D(P*GP*GP*AP*AP*TP*TP*AP*TP*AP*AP*TP*TP*CP*C)-3'), Interferon-activable protein 202 | | Authors: | Ru, H, Ni, X, Crowley, C, Zhao, L, Ding, W, Hung, L.-W, Shaw, N, Cheng, G, Liu, Z.-J. | | Deposit date: | 2013-02-19 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.963 Å) | | Cite: | Structural basis for termination of AIM2-mediated signaling by p202
Cell Res., 23, 2013
|
|
4JBM
 
 | | Structure of murine DNA binding protein bound with ds DNA | | Descriptor: | DNA (5'-D(*GP*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP*C)-3'), Interferon-inducible protein AIM2 | | Authors: | Ru, H, Ni, X, Crowley, C, Zhao, L, Ding, W, Hung, L.-W, Shaw, N, Cheng, G, Liu, Z.-J. | | Deposit date: | 2013-02-19 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.218 Å) | | Cite: | Structural basis for termination of AIM2-mediated signaling by p202
Cell Res., 23, 2013
|
|
6DBR
 
 | | Cryo-EM structure of RAG in complex with one melted RSS and one unmelted RSS | | Descriptor: | CALCIUM ION, Forward strand of melted RSS substrate DNA, Forward strand of unmelted RSS substrate DNA, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBL
 
 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | Descriptor: | CALCIUM ION, Molecule name: Forward strand of 12-RSS substrate DNA, Molecule name: Forward strand of 23-RSS substrate DNA, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (5.001 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBQ
 
 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | Descriptor: | CALCIUM ION, Molecule name: Forward strand of 12-RSS substrate DNA, Molecule name: Forward strand of 23-RSS substrate DNA, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.22 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBW
 
 | | Cryo-EM structure of RAG in complex with 12-RSS substrate DNA | | Descriptor: | CALCIUM ION, Forward strand of 12-RSS substrate DNA, Recombination activating gene 1 - MBP chimera, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBT
 
 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | Descriptor: | CALCIUM ION, Forward strand of 12-RSS substrate DNA, Forward strand of 23-RSS substrate DNA, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBX
 
 | | Cryo-EM structure of RAG in complex with 12-RSS substrate DNA | | Descriptor: | CALCIUM ION, Forward strand of 12-RSS substrate DNA, Recombination activating gene 1 - MBP chimera, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBU
 
 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | Descriptor: | CALCIUM ION, Forward strand RSS substrate DNA, Recombination activating gene 1 - MBP chimera, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBI
 
 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS nicked DNA intermediates | | Descriptor: | CALCIUM ION, Forward strand of 12-RSS signal end, Forward strand of 23-RSS signal end, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBJ
 
 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS nicked DNA intermediates | | Descriptor: | CALCIUM ION, Forward stand of RSS signal end, Forward strand of coding flank, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBO
 
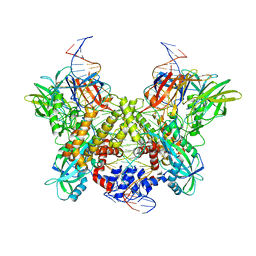 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | Descriptor: | CALCIUM ION, Forward strand of substrate RSS DNA, Recombination activating gene 1 - MBP chimera, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBV
 
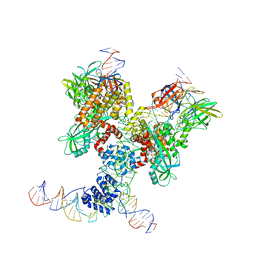 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | Descriptor: | CALCIUM ION, Forward strand of 12-RSS substrate DNA, Forward strand of 23-RSS substrate DNA, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.291 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
4EF4
 
 | | Crystal structure of STING CTD complex with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CALCIUM ION, Transmembrane protein 173 | | Authors: | Ouyang, S, Ru, H, Shaw, N, Jiang, Y, Niu, F, Zhu, Y, Qiu, W, Li, Y, Liu, Z.-J. | | Deposit date: | 2012-03-29 | | Release date: | 2012-05-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding
Immunity, 36, 2012
|
|
4EF5
 
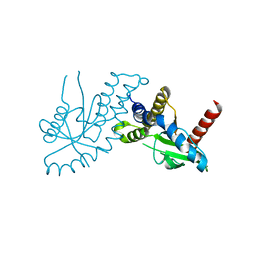 | | Crystal structure of STING CTD | | Descriptor: | Transmembrane protein 173 | | Authors: | Ouyang, S, Ru, H, Shaw, N, Jiang, Y, Niu, F, Zhu, Y, Qiu, W, Li, Y, Liu, Z.-J. | | Deposit date: | 2012-03-29 | | Release date: | 2012-05-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding
Immunity, 36, 2012
|
|
6DLO
 
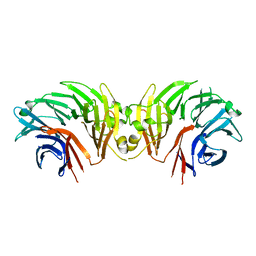 | |
