7PQH
 
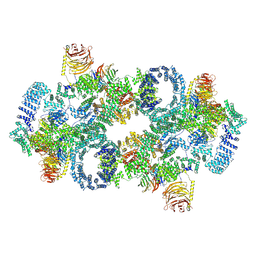 | | Cryo-EM structure of Saccharomyces cerevisiae TOROID (TORC1 Organized in Inhibited Domains). | | Descriptor: | Serine/threonine-protein kinase TOR2, Target of rapamycin complex 1 subunit KOG1,Target of rapamycin complex 1 subunit Kog1, Target of rapamycin complex subunit LST8 | | Authors: | Felix, J, Prouteau, M, Bourgoint, C, Bonadei, L, Desfosses, A, Gabus, C, Sadian, Y, Savvides, S.N, Gutsche, I, Loewith, R. | | Deposit date: | 2021-09-17 | | Release date: | 2023-01-18 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | EGOC inhibits TOROID polymerization by structurally activating TORC1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6EMK
 
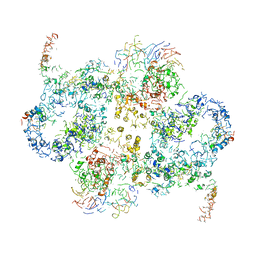 | | Cryo-EM Structure of Saccharomyces cerevisiae Target of Rapamycin Complex 2 | | Descriptor: | Serine/threonine-protein kinase TOR2, Target of rapamycin complex 2 subunit AVO1, Target of rapamycin complex 2 subunit AVO2, ... | | Authors: | Karuppasamy, M, Kusmider, B, Oliveira, T.M, Gaubitz, C, Prouteau, M, Loewith, R, Schaffitzel, C. | | Deposit date: | 2017-10-02 | | Release date: | 2017-12-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Cryo-EM structure of Saccharomyces cerevisiae target of rapamycin complex 2.
Nat Commun, 8, 2017
|
|
8S86
 
 | | human PLD3 homodimer structure | | Descriptor: | 5'-3' exonuclease PLD3 | | Authors: | Lammens, K. | | Deposit date: | 2024-03-05 | | Release date: | 2024-05-15 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Lysosomal endonuclease RNase T2 and PLD exonucleases cooperatively generate RNA ligands for TLR7 activation.
Immunity, 57, 2024
|
|
3KFO
 
 | | Crystal structure of the C-terminal domain from the nuclear pore complex component NUP133 from Saccharomyces cerevisiae | | Descriptor: | GLYCEROL, Nucleoporin NUP133 | | Authors: | Sampathkumar, P, Bonanno, J.B, Miller, S, Bain, K, Dickey, M, Gheyi, T, Almo, S.C, Rout, M, Sali, A, Phillips, J, Pieper, U, Fernandez-Martinez, J, Franke, J.D, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-27 | | Release date: | 2010-01-26 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the C-terminal domain of Saccharomyces cerevisiae Nup133, a component of the nuclear pore complex.
Proteins, 79, 2011
|
|
3KES
 
 | | Crystal structure of the autoproteolytic domain from the nuclear pore complex component NUP145 from Saccharomyces cerevisiae in the Hexagonal, P61 space group | | Descriptor: | 1,2-ETHANEDIOL, Nucleoporin NUP145 | | Authors: | Sampathkumar, P, Ozyurt, S.A, Do, J, Bain, K, Dickey, M, Gheyi, T, Sali, A, Kim, S.J, Phillips, J, Pieper, U, Fernandez-Martinez, J, Franke, J.D, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Rout, M, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-26 | | Release date: | 2009-12-22 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the autoproteolytic domain from the Saccharomyces cerevisiae nuclear pore complex component, Nup145.
Proteins, 78, 2010
|
|
3KEP
 
 | | Crystal structure of the autoproteolytic domain from the nuclear pore complex component NUP145 from Saccharomyces cerevisiae | | Descriptor: | 1,2-ETHANEDIOL, Nucleoporin NUP145 | | Authors: | Sampathkumar, P, Ozyurt, S.A, Do, J, Bain, K, Dickey, M, Gheyi, T, Sali, A, Kim, S.J, Phillips, J, Pieper, U, Fernandez-Martinez, J, Franke, J.D, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Rout, M, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-26 | | Release date: | 2009-12-22 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structures of the autoproteolytic domain from the Saccharomyces cerevisiae nuclear pore complex component, Nup145.
Proteins, 78, 2010
|
|
3NF5
 
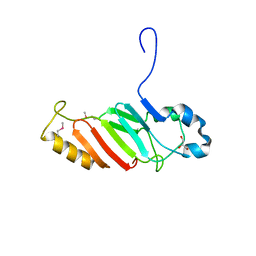 | | Crystal structure of the C-terminal domain of nuclear pore complex component NUP116 from Candida glabrata | | Descriptor: | GLYCEROL, Nucleoporin NUP116 | | Authors: | Sampathkumar, P, Manglicmot, D, Bain, K, Gilmore, J, Gheyi, T, Rout, M, Sali, A, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-09 | | Release date: | 2010-08-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Atomic structure of the nuclear pore complex targeting domain of a Nup116 homologue from the yeast, Candida glabrata.
Proteins, 80, 2012
|
|
3CQU
 
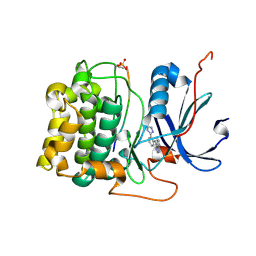 | | Crystal Structure of Akt-1 complexed with substrate peptide and inhibitor | | Descriptor: | Glycogen synthase kinase-3 beta, N-[2-(5-methyl-4H-1,2,4-triazol-3-yl)phenyl]-7H-pyrrolo[2,3-d]pyrimidin-4-amine, RAC-alpha serine/threonine-protein kinase | | Authors: | Pandit, J. | | Deposit date: | 2008-04-03 | | Release date: | 2008-05-27 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synthesis and structure based optimization of novel Akt inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3CQW
 
 | | Crystal Structure of Akt-1 complexed with substrate peptide and inhibitor | | Descriptor: | 5-(5-chloro-7H-pyrrolo[2,3-d]pyrimidin-4-yl)-4,5,6,7-tetrahydro-1H-imidazo[4,5-c]pyridine, Glycogen synthase kinase-3 beta, MANGANESE (II) ION, ... | | Authors: | Pandit, J. | | Deposit date: | 2008-04-03 | | Release date: | 2008-05-27 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis and structure based optimization of novel Akt inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3UAU
 
 | | Crystal structure of the lipoprotein JlpA | | Descriptor: | Surface-exposed lipoprotein | | Authors: | Kawai, F, Yeo, H.J. | | Deposit date: | 2011-10-22 | | Release date: | 2012-07-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of JlpA, a surface-exposed lipoprotein adhesin of Campylobacter jejuni.
J.Struct.Biol., 177, 2012
|
|
