4DAA
 
 | |
3DAA
 
 | |
2DAA
 
 | |
1NW1
 
 | | Crystal Structure of Choline Kinase | | Descriptor: | CALCIUM ION, Choline kinase (49.2 kD) | | Authors: | Peisach, D, Gee, P, Kent, C, Xu, Z. | | Deposit date: | 2003-02-05 | | Release date: | 2003-06-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The Crystal Structure of Choline Kinase Reveals a Eukaryotic Protein Kinase Fold
Structure, 11, 2003
|
|
5DAA
 
 | |
1A0G
 
 | | L201A MUTANT OF D-AMINO ACID AMINOTRANSFERASE COMPLEXED WITH PYRIDOXAMINE-5'-PHOSPHATE | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, D-AMINO ACID AMINOTRANSFERASE | | Authors: | Sugio, S, Kashima, A, Kishimoto, K, Peisach, D, Petsko, G.A, Ringe, D, Yoshimura, T, Esaki, N. | | Deposit date: | 1997-11-30 | | Release date: | 1998-06-03 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of L201A mutant of D-amino acid aminotransferase at 2.0 A resolution: implication of the structural role of Leu201 in transamination.
Protein Eng., 11, 1998
|
|
4RHN
 
 | | HISTIDINE TRIAD NUCLEOTIDE-BINDING PROTEIN (HINT) FROM RABBIT COMPLEXED WITH ADENOSINE | | Descriptor: | HISTIDINE TRIAD NUCLEOTIDE-BINDING PROTEIN, alpha-D-ribofuranose | | Authors: | Brenner, C, Garrison, P, Gilmour, J, Peisach, D, Ringe, D, Petsko, G.A, Lowenstein, J.M. | | Deposit date: | 1997-02-26 | | Release date: | 1997-06-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of HINT demonstrate that histidine triad proteins are GalT-related nucleotide-binding proteins.
Nat.Struct.Biol., 4, 1997
|
|
6RHN
 
 | | HISTIDINE TRIAD NUCLEOTIDE-BINDING PROTEIN (HINT) FROM RABBIT WITHOUT NUCLEOTIDE | | Descriptor: | HISTIDINE TRIAD NUCLEOTIDE-BINDING PROTEIN | | Authors: | Brenner, C, Garrison, P, Gilmour, J, Peisach, D, Ringe, D, Petsko, G.A, Lowenstein, J.M. | | Deposit date: | 1997-02-27 | | Release date: | 1997-06-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of HINT demonstrate that histidine triad proteins are GalT-related nucleotide-binding proteins.
Nat.Struct.Biol., 4, 1997
|
|
3RHN
 
 | | HISTIDINE TRIAD NUCLEOTIDE-BINDING PROTEIN (HINT) FROM RABBIT COMPLEXED WITH GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, HISTIDINE TRIAD NUCLEOTIDE-BINDING PROTEIN | | Authors: | Brenner, C, Garrison, P, Gilmour, J, Peisach, D, Ringe, D, Petsko, G.A, Lowenstein, J.M. | | Deposit date: | 1997-02-11 | | Release date: | 1997-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of HINT demonstrate that histidine triad proteins are GalT-related nucleotide-binding proteins.
Nat.Struct.Biol., 4, 1997
|
|
5RHN
 
 | | HISTIDINE TRIAD NUCLEOTIDE-BINDING PROTEIN (HINT) FROM RABBIT COMPLEXED WITH 8-BR-AMP | | Descriptor: | 8-BROMO-ADENOSINE-5'-MONOPHOSPHATE, HISTIDINE TRIAD NUCLEOTIDE-BINDING PROTEIN | | Authors: | Brenner, C, Garrison, P, Gilmour, J, Peisach, D, Ringe, D, Petsko, G.A, Lowenstein, J.M. | | Deposit date: | 1997-02-26 | | Release date: | 1997-06-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal structures of HINT demonstrate that histidine triad proteins are GalT-related nucleotide-binding proteins.
Nat.Struct.Biol., 4, 1997
|
|
2DAB
 
 | | L201A MUTANT OF D-AMINO ACID AMINOTRANSFERASE COMPLEXED WITH PYRIDOXAL-5'-PHOSPHATE | | Descriptor: | D-AMINO ACID AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Sugio, S, Kashima, A, Kishimoto, K, Peisach, D, Petsko, G.A, Ringe, D, Yoshimura, T, Esaki, N. | | Deposit date: | 1997-11-30 | | Release date: | 1998-06-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of L201A mutant of D-amino acid aminotransferase at 2.0 A resolution: implication of the structural role of Leu201 in transamination.
Protein Eng., 11, 1998
|
|
1ZCP
 
 | |
1R62
 
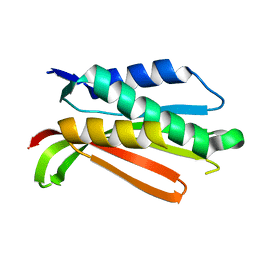 | | Crystal structure of the C-terminal Domain of the Two-Component System Transmitter Protein NRII (NtrB) | | Descriptor: | Nitrogen regulation protein NR(II) | | Authors: | Song, Y, Peisach, D, Pioszak, A.A, Xu, Z, Ninfa, A.J. | | Deposit date: | 2003-10-14 | | Release date: | 2004-06-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the C-terminal Domain of the Two-Component System Transmitter Protein Nitrogen Regulator II (NRII; NtrB), Regulator of Nitrogen Assimilation in Escherichia coli.
Biochemistry, 43, 2004
|
|
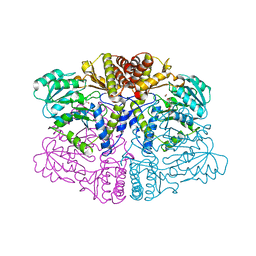
6PK3
 
 | |
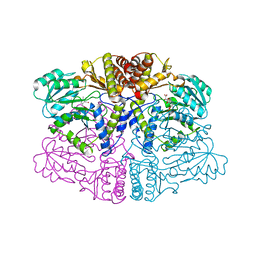
6PK1
 
 | |
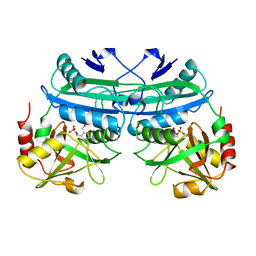
1DAA
 
 | |
1G2W
 
 | | E177S MUTANT OF THE PYRIDOXAL-5'-PHOSPHATE ENZYME D-AMINO ACID AMINOTRANSFERASE | | Descriptor: | ACETATE ION, D-ALANINE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Lepore, B.W, Ringe, D. | | Deposit date: | 2000-10-21 | | Release date: | 2000-11-15 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Studies on an Active Site Residue, E177, That Affects Binding of the Coenzyme in D-Amino Acid Transaminase, and Mechanistic Studies on a Suicide Substrate
Biochemistry and Molecular Biology of Vitamin B6 and PQQ-dependent Proteins, 10th Annual International Symposium on Vitamin B6, 2000
|
|
2HZ6
 
 | | The crystal structure of human IRE1-alpha luminal domain | | Descriptor: | Endoplasmic reticulum to nucleus signalling 1 isoform 1 variant | | Authors: | Kaufman, R.J, Xu, Z, Zhou, J. | | Deposit date: | 2006-08-08 | | Release date: | 2006-08-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The crystal structure of human IRE1 luminal domain reveals a conserved dimerization interface required for activation of the unfolded protein response.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1RZY
 
 | | Crystal structure of rabbit Hint complexed with N-ethylsulfamoyladenosine | | Descriptor: | 5'-O-(N-ETHYL-SULFAMOYL)ADENOSINE, Histidine triad nucleotide-binding protein 1 | | Authors: | Krakowiak, A.K, Pace, H.C, Blackburn, G.M, Adams, M, Mekhalfia, A, Kaczmarek, R, Baraniak, J, Stec, W.J, Brenner, C. | | Deposit date: | 2003-12-29 | | Release date: | 2004-03-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biochemical, crystallographic, and mutagenic characterization of hint, the AMP-lysine hydrolase, with novel substrates and inhibitors
J.Biol.Chem., 279, 2004
|
|
