6IDG
 
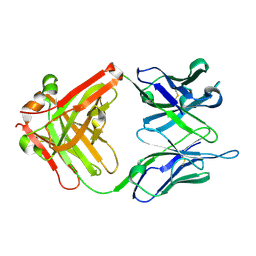 | | antibody 64M-5 Fab in complex with dT(6-4)T | | Descriptor: | Anti-(6-4) photoproduct antibody 64M-5 Fab (heavy chain), Anti-(6-4) photoproduct antibody 64M-5 Fab (light chain), DNA (5'-D(*(64T)P*(5PY))-3') | | Authors: | Yokoyama, H, Mizutani, R, Noguchi, S, Hayashida, N. | | Deposit date: | 2018-09-10 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the antibody 64M-5 Fab and its complex with dT(6-4)T indicate induced-fit and high-affinity mechanisms.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
4EVD
 
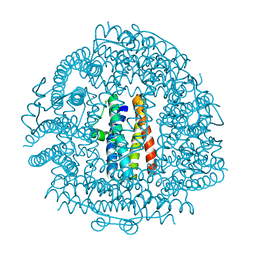 | | Crystal Structure HP-NAP from strain YS29 cadmium loaded (Cocrystallization 50mM) | | Descriptor: | CADMIUM ION, Neutrophil-activating protein | | Authors: | Yokoyama, H, Tsuruta, O, Akao, N, Fujii, S. | | Deposit date: | 2012-04-26 | | Release date: | 2012-06-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Helicobacter pylori neutrophil-activating protein with a di-nuclear ferroxidase center in a zinc or cadmium-bound form
Biochem.Biophys.Res.Commun., 422, 2012
|
|
4EVE
 
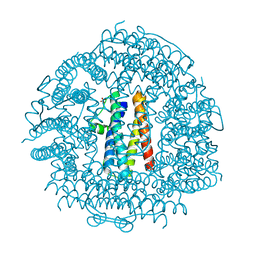 | | Crystal Structure HP-NAP from strain YS29 in apo form | | Descriptor: | Neutrophil-activating protein, SULFATE ION | | Authors: | Yokoyama, H, Tsuruta, O, Akao, N, Fujii, S. | | Deposit date: | 2012-04-26 | | Release date: | 2012-06-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Helicobacter pylori neutrophil-activating protein with a di-nuclear ferroxidase center in a zinc or cadmium-bound form
Biochem.Biophys.Res.Commun., 422, 2012
|
|
4EVC
 
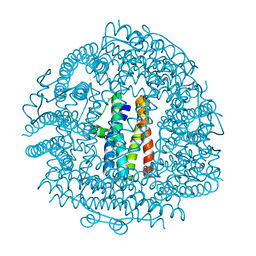 | | Crystal Structure HP-NAP from strain YS39 cadmium loaded (Cocrystallization 50mM) | | Descriptor: | CADMIUM ION, Neutrophil-activating protein | | Authors: | Yokoyama, H, Tsuruta, O, Akao, N, Fujii, S. | | Deposit date: | 2012-04-26 | | Release date: | 2012-06-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Helicobacter pylori neutrophil-activating protein with a di-nuclear ferroxidase center in a zinc or cadmium-bound form
Biochem.Biophys.Res.Commun., 422, 2012
|
|
4EVB
 
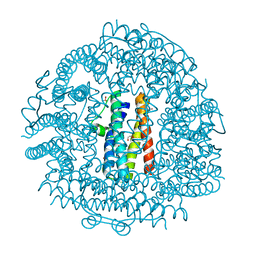 | | Crystal Structure HP-NAP from strain YS39 zinc soaked (20mM) | | Descriptor: | 1,2-ETHANEDIOL, Neutrophil-activating protein, SULFATE ION, ... | | Authors: | Yokoyama, H, Tsuruta, O, Akao, N, Fujii, S. | | Deposit date: | 2012-04-26 | | Release date: | 2012-06-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Helicobacter pylori neutrophil-activating protein with a di-nuclear ferroxidase center in a zinc or cadmium-bound form
Biochem.Biophys.Res.Commun., 422, 2012
|
|
3BK6
 
 | |
1EHL
 
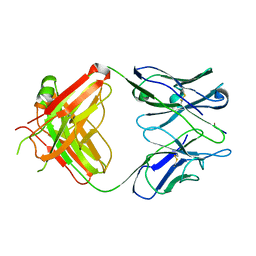 | | 64M-2 ANTIBODY FAB COMPLEXED WITH D(5HT)(6-4)T | | Descriptor: | 5'-(D(5HT)P*(6-4)T)-3', ANTI-(6-4) PHOTOPRODUCT ANTIBODY 64M-2 FAB (HEAVY CHAIN), ANTI-(6-4) PHOTOPRODUCT ANTIBODY 64M-2 FAB (LIGHT CHAIN) | | Authors: | Yokoyama, H, Mizutani, R, Satow, Y, Komatsu, Y, Ohtsuka, E, Nikaido, O. | | Deposit date: | 2000-02-21 | | Release date: | 2001-02-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the 64M-2 antibody Fab fragment in complex with a DNA dT(6-4)T photoproduct formed by ultraviolet radiation.
J.Mol.Biol., 299, 2000
|
|
3VIV
 
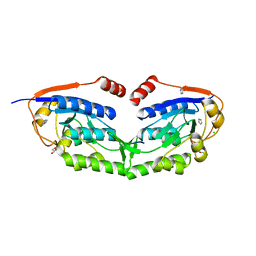 | | 1510-N membrane-bound stomatin-specific protease K138A mutant in complex with a substrate peptide | | Descriptor: | 441aa long hypothetical nfeD protein, CHLORIDE ION, GLYCEROL, ... | | Authors: | Yokoyama, H, Matsui, I, Fujii, S. | | Deposit date: | 2011-10-12 | | Release date: | 2012-05-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of a membrane stomatin-specific protease in complex with a substrate Peptide
Biochemistry, 51, 2012
|
|
3VW3
 
 | | Antibody 64M-5 Fab in complex with a double-stranded DNA (6-4) photoproduct | | Descriptor: | Anti-(6-4) photoproduct antibody 64M-5 Fab (heavy chain), Anti-(6-4) photoproduct antibody 64M-5 Fab (light chain), COBALT HEXAMMINE(III), ... | | Authors: | Yokoyama, H, Mizutani, R, Satow, Y. | | Deposit date: | 2012-07-30 | | Release date: | 2013-03-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a double-stranded DNA (6-4) photoproduct in complex with the 64M-5 antibody Fab
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3WG5
 
 | | 1510-N membrane-bound stomatin-specific protease K138A mutant in complex with a substrate peptide under heat treatment | | Descriptor: | 441aa long hypothetical nfeD protein, CHLORIDE ION, GLYCEROL, ... | | Authors: | Yokoyama, H, Fujii, S, Matsui, I. | | Deposit date: | 2013-07-26 | | Release date: | 2013-10-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and biochemical analysis of a thermostable membrane-bound stomatin-specific protease.
J.Synchrotron Radiat., 20, 2013
|
|
3WWV
 
 | |
6IDH
 
 | | Antibody 64M-5 Fab in ligand-free form | | Descriptor: | Anti-(6-4) photoproduct antibody 64M-5 Fab (heavy chain), Anti-(6-4) photoproduct antibody 64M-5 Fab (light chain) | | Authors: | Yokoyama, H, Mizutani, R, Noguchi, S, Hayashida, N. | | Deposit date: | 2018-09-10 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the antibody 64M-5 Fab and its complex with dT(6-4)T indicate induced-fit and high-affinity mechanisms.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6M4B
 
 | |
1KEG
 
 | | Antibody 64M-2 Fab complexed with dTT(6-4)TT | | Descriptor: | 5'-D(*TP*(64T)P*TP*T)-3', Anti-(6-4) photoproduct antibody 64M-2 Fab (heavy chain), Anti-(6-4) photoproduct antibody 64M-2 Fab (light chain), ... | | Authors: | Yokoyama, H, Mizutani, R, Satow, Y, Sato, K, Komatsu, Y, Ohtsuka, E, Nikaido, O. | | Deposit date: | 2001-11-15 | | Release date: | 2002-11-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the DNA (6-4) photoproduct dTT(6-4)TT in complex with the 64M-2 antibody Fab fragment implies increased antibody-binding affinity by the flanking nucleotides.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3WPN
 
 | |
7FI3
 
 | | Archaeal oligopeptide permease A (OppA) from Thermococcus kodakaraensis in complex with an endogenous pentapeptide | | Descriptor: | ABC-type dipeptide/oligopeptide transport system, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Yokoyama, H, Kamei, N, Konishi, K, Hara, K, Hashimoto, H. | | Deposit date: | 2021-07-30 | | Release date: | 2022-04-13 | | Last modified: | 2022-06-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for peptide recognition by archaeal oligopeptide permease A.
Proteins, 90, 2022
|
|
6KDI
 
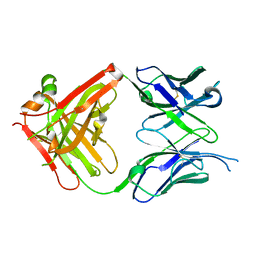 | | Antibody 64M-5 Fab including isoAsp in complex with dT(6-4)T | | Descriptor: | Anti-(6-4) photoproduct antibody 64M-5 Fab (heavy chain), Anti-(6-4) photoproduct antibody 64M-5 Fab (light chain), DNA (5'-D(*(64T)P*(5PY))-3') | | Authors: | Yokoyama, H, Mizutani, R, Noguchi, S, Hayashida, N. | | Deposit date: | 2019-07-02 | | Release date: | 2019-12-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and biochemical basis of the formation of isoaspartate in the complementarity-determining region of antibody 64M-5 Fab.
Sci Rep, 9, 2019
|
|
6KDH
 
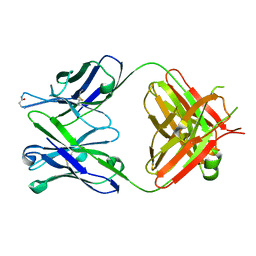 | | Antibody 64M-5 Fab including isoAsp in ligand-free form | | Descriptor: | Anti-(6-4) photoproduct antibody 64M-5 Fab (heavy chain), Anti-(6-4) photoproduct antibody 64M-5 Fab (light chain) | | Authors: | Yokoyama, H, Mizutani, R, Noguchi, S, Hayashida, N. | | Deposit date: | 2019-07-02 | | Release date: | 2019-12-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural and biochemical basis of the formation of isoaspartate in the complementarity-determining region of antibody 64M-5 Fab.
Sci Rep, 9, 2019
|
|
5ZO8
 
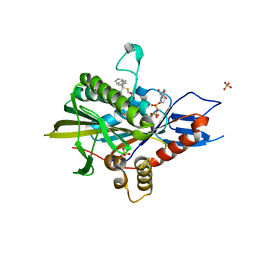 | | Eg5 motor domain in complex with STLC-type inhibitor PVEI0021 (P21 type) | | Descriptor: | (2R)-2-azanyl-3-[(4-methoxyphenyl)-diphenyl-methyl]sulfanyl-propanoic acid, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Yokoyama, H, Sato, K. | | Deposit date: | 2018-04-12 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Thermodynamic Basis of the Enhanced Interaction between Kinesin Spindle Protein Eg5 and STLC-type Inhibitors.
Acs Omega, 3, 2018
|
|
5ZO9
 
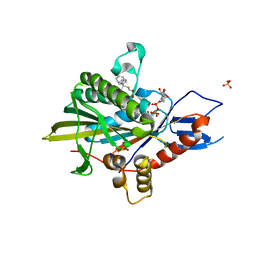 | | Eg5 motor domain in complex with STLC-type inhibitor PVEI0021 (C2 type) | | Descriptor: | (2R)-2-azanyl-3-[(4-methoxyphenyl)-diphenyl-methyl]sulfanyl-propanoic acid, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Yokoyama, H, Sato, K. | | Deposit date: | 2018-04-12 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Thermodynamic Basis of the Enhanced Interaction between Kinesin Spindle Protein Eg5 and STLC-type Inhibitors.
Acs Omega, 3, 2018
|
|
1YIQ
 
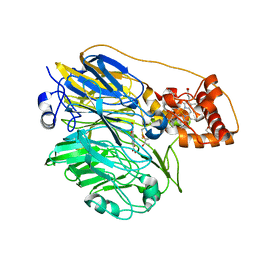 | | Molecular cloning and structural analysis of quinohemoprotein alcohol dehydrogenase ADHIIG from Pseudomonas putida HK5. Compariison to the other quinohemoprotein alcohol dehydrogenase ADHIIB found in the same microorganism. | | Descriptor: | CALCIUM ION, HEME C, PYRROLOQUINOLINE QUINONE, ... | | Authors: | Toyama, H, Chen, Z.W, Fukumoto, M, Adachi, O, Matsushita, K, Mathews, F.S. | | Deposit date: | 2005-01-12 | | Release date: | 2005-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular cloning and structural analysis of quinohemoprotein alcohol dehydrogenase ADH-IIG from Pseudomonas putida HK5
J.Mol.Biol., 352, 2005
|
|
5ZO7
 
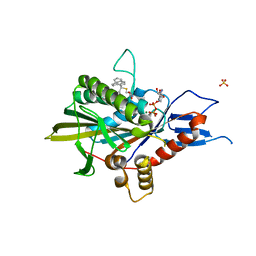 | | Kinesin spindle protein Eg5 in complex with STLC-type inhibitor PVEI0138 | | Descriptor: | (2R)-2-azanyl-3-[[2-(4-methoxyphenyl)-2-tricyclo[9.4.0.0^{3,8}]pentadeca-1(11),3,5,7,12,14-hexaenyl]sulfanyl]propanoic acid, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Yokoyama, H, Sato, K. | | Deposit date: | 2018-04-12 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Thermodynamic Basis of the Enhanced Interaction between Kinesin Spindle Protein Eg5 and STLC-type Inhibitors.
Acs Omega, 3, 2018
|
|
7WXS
 
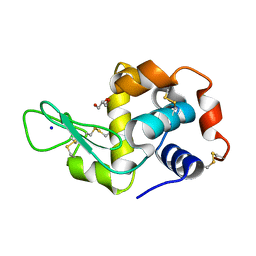 | | Lysozyme protected by polyacrylamide gel | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme C, ... | | Authors: | Muroyama, H, Tomoike, F, Nagae, T, Okada, T. | | Deposit date: | 2022-02-15 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Post-crystallization protection of protein crystals by polyacrylamide
To Be Published
|
|
7WXT
 
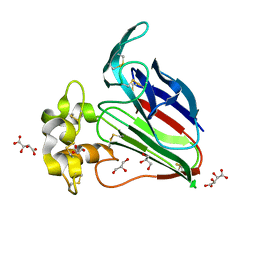 | |
3BPP
 
 | |
