6XFK
 
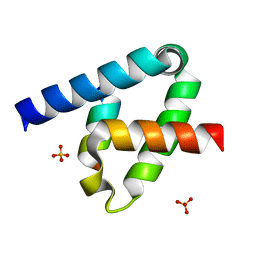 | | Crystal structure of the type III secretion system pilotin-secretin complex InvH-InvG | | Descriptor: | SULFATE ION, Type 3 secretion system pilotin, Type 3 secretion system secretin | | Authors: | Majewski, D.D, Okon, M, Heinkel, F, Robb, C.S, Vuckovic, M, McIntosh, L.P, Strynadka, N.C.J. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Characterization of the Pilotin-Secretin Complex from the Salmonella enterica Type III Secretion System Using Hybrid Structural Methods.
Structure, 29, 2021
|
|
6XFJ
 
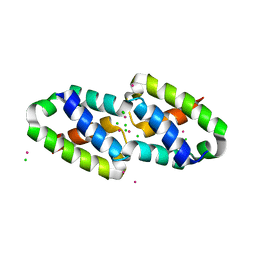 | | Crystal structure of the type III secretion pilotin InvH | | Descriptor: | CADMIUM ION, CHLORIDE ION, SODIUM ION, ... | | Authors: | Majewski, D.D, Okon, M, Heinkel, F, Robb, C.S, Vuckovic, M, McIntosh, L.P, Strynadka, N.C.J. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Characterization of the Pilotin-Secretin Complex from the Salmonella enterica Type III Secretion System Using Hybrid Structural Methods.
Structure, 29, 2021
|
|
6XFL
 
 | | Structural characterization of the type III secretion system pilotin-secretin complex InvH-InvG by NMR spectroscopy | | Descriptor: | Type 3 secretion system pilotin, Type 3 secretion system secretin | | Authors: | Majewski, D.D, Okon, M, Heinkel, F, Robb, C.S, Vuckovic, M, McIntosh, L.P, Strynadka, N.C.J. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Characterization of the Pilotin-Secretin Complex from the Salmonella enterica Type III Secretion System Using Hybrid Structural Methods.
Structure, 29, 2021
|
|
4MHG
 
 | | Crystal structure of ETV6 bound to a specific DNA sequence | | Descriptor: | Complementary Specific 14 bp DNA, Specific 14 bp DNA, Transcription factor ETV6 | | Authors: | Chan, A.C, De, S, Coyne III, H.J, Okon, M, Murphy, M.E, Graves, B.J, McIntosh, L.P. | | Deposit date: | 2013-08-29 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Steric Mechanism of Auto-Inhibitory Regulation of Specific and Non-Specific DNA Binding by the ETS Transcriptional Repressor ETV6.
J.Mol.Biol., 426, 2014
|
|
1T5N
 
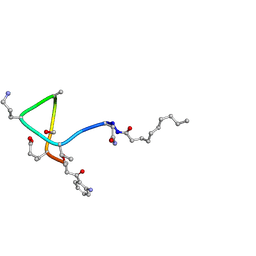 | | Structural transitions as determinants of calcium-dependent antibiotic daptomycin | | Descriptor: | DAPTOMYCIN, DECANOIC ACID | | Authors: | Jung, D, Rozek, A, Okon, M, Hancock, R.E. | | Deposit date: | 2004-05-04 | | Release date: | 2004-08-31 | | Last modified: | 2025-03-26 | | Method: | SOLUTION NMR | | Cite: | Structural Transitions as Determinants of the Action of the Calcium-Dependent Antibiotic Daptomycin.
Chem.Biol., 11, 2004
|
|
1T5M
 
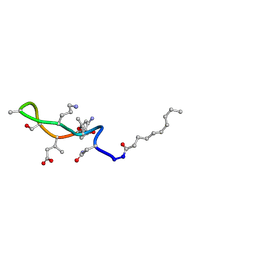 | | Structural transitions as determinants of the action of the calcium-dependent antibiotic daptomycin | | Descriptor: | DAPTOMYCIN, DECANOIC ACID | | Authors: | Jung, D, Rozek, A, Okon, M, Hancock, R.E. | | Deposit date: | 2004-05-04 | | Release date: | 2004-08-31 | | Last modified: | 2025-03-26 | | Method: | SOLUTION NMR | | Cite: | Structural Transitions as Determinants of the Action of the Calcium-Dependent Antibiotic Daptomycin.
Chem.Biol., 11, 2004
|
|
6CAH
 
 | |
6DA1
 
 | | ETS1 in complex with synthetic SRR mimic | | Descriptor: | Protein C-ets-1, SULFATE ION, serine-rich region (SRR) peptide | | Authors: | Perez-Borrajero, C, Okon, M, Lin, C.S, Scheu, K, Murphy, M.E.P, Graves, B.J, McIntosh, L.P. | | Deposit date: | 2018-05-01 | | Release date: | 2019-01-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.000127 Å) | | Cite: | The Biophysical Basis for Phosphorylation-Enhanced DNA-Binding Autoinhibition of the ETS1 Transcription Factor.
J. Mol. Biol., 431, 2019
|
|
6DAT
 
 | | ETS1 in complex with synthetic SRR mimic | | Descriptor: | Protein C-ets-1, SULFATE ION, serine-rich region (SRR) peptide | | Authors: | Perez-Borrajero, C, Okon, M, Lin, C.S, Scheu, K, Murphy, M.E.P, Graves, B.J, McIntosh, L.P. | | Deposit date: | 2018-05-02 | | Release date: | 2019-01-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.35002637 Å) | | Cite: | The Biophysical Basis for Phosphorylation-Enhanced DNA-Binding Autoinhibition of the ETS1 Transcription Factor.
J. Mol. Biol., 431, 2019
|
|
2LV4
 
 | | ZirS C-terminal Domain | | Descriptor: | Putative outer membrane or exported protein | | Authors: | Prehna, G, Li, Y, Stoynov, N, Okon, M, Vukovic, M, Mcintosh, L.P, Foster, L.J, Finlay, B, Strynadka, N.C.J. | | Deposit date: | 2012-06-28 | | Release date: | 2012-08-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The zinc regulated antivirulence pathway of salmonella is a multiprotein immunoglobulin adhesion system.
J.Biol.Chem., 287, 2012
|
|
3VZL
 
 | | Crystal structure of the Bacillus circulans endo-beta-(1,4)-xylanase (BcX) N35H mutant | | Descriptor: | Endo-1,4-beta-xylanase, SULFATE ION | | Authors: | Ludwiczek, M.L, D'Angelo, I, Yalloway, G.N, Okon, M, Nielsen, J.E, Strynadka, N.C, Withers, S.G, McIntosh, L.P. | | Deposit date: | 2012-10-15 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Strategies for modulating the pH-dependent activity of a family 11 glycoside hydrolase
Biochemistry, 52, 2013
|
|
3VZN
 
 | | Crystal structure of the Bacillus circulans endo-beta-(1,4)-xylanase (BcX) N35E mutant with Glu78 covalently bonded to 2-deoxy-2-fluoro-xylobiose | | Descriptor: | Endo-1,4-beta-xylanase, SULFATE ION, beta-D-xylopyranose-(1-4)-1,5-anhydro-2-deoxy-2-fluoro-D-xylitol | | Authors: | Ludwiczek, M.L, D'Angelo, I, Yalloway, G.N, Okon, M, Nielsen, J.E, Strynadka, N.C, Withers, S.G, McIntosh, L.P. | | Deposit date: | 2012-10-15 | | Release date: | 2013-05-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Strategies for modulating the pH-dependent activity of a family 11 glycoside hydrolase
Biochemistry, 52, 2013
|
|
3VZJ
 
 | | Crystal structure of the Bacillus circulans endo-beta-(1,4)-xylanase (BcX) E172H mutant | | Descriptor: | Endo-1,4-beta-xylanase, SULFATE ION | | Authors: | Ludwiczek, M.L, D'Angelo, I, Yalloway, G.N, Okon, M, Nielsen, J.E, Strynadka, N.C, Withers, S.G, McIntosh, L.P. | | Deposit date: | 2012-10-14 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Strategies for modulating the pH-dependent activity of a family 11 glycoside hydrolase
Biochemistry, 52, 2013
|
|
3VZM
 
 | | Crystal structure of the Bacillus circulans endo-beta-(1,4)-xylanase (BcX) E172H mutant with Glu78 covalently bonded to 2-deoxy-2-fluoro-xylobiose | | Descriptor: | Endo-1,4-beta-xylanase, beta-D-xylopyranose-(1-4)-1,5-anhydro-2-deoxy-2-fluoro-D-xylitol | | Authors: | Ludwiczek, M.L, D'Angelo, I, Yalloway, G.N, Okon, M, Nielsen, J.E, Strynadka, N.C, Withers, S.G, McIntosh, L.P. | | Deposit date: | 2012-10-15 | | Release date: | 2013-05-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Strategies for modulating the pH-dependent activity of a family 11 glycoside hydrolase
Biochemistry, 52, 2013
|
|
3VZK
 
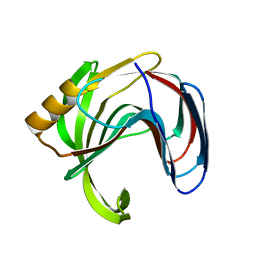 | | Crystal structure of the Bacillus circulans endo-beta-(1,4)-xylanase (BcX) N35E mutant | | Descriptor: | Endo-1,4-beta-xylanase, SULFATE ION | | Authors: | Ludwiczek, M.L, D'Angelo, I, Yalloway, G.N, Okon, M, Nielsen, J.E, Strynadka, N.C, Withers, S.G, McIntosh, L.P. | | Deposit date: | 2012-10-14 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Strategies for modulating the pH-dependent activity of a family 11 glycoside hydrolase
Biochemistry, 52, 2013
|
|
3VZO
 
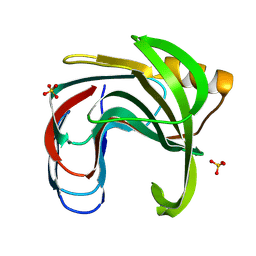 | | Crystal structure of the Bacillus circulans endo-beta-(1,4)-xylanase (BcX) N35H mutant with Glu78 covalently bonded to 2-deoxy-2-fluoro-xylobiose | | Descriptor: | Endo-1,4-beta-xylanase, SULFATE ION, beta-D-xylopyranose-(1-4)-1,5-anhydro-2-deoxy-2-fluoro-D-xylitol | | Authors: | Ludwiczek, M.L, D'Angelo, I, Yalloway, G.N, Okon, M, Nielsen, J.E, Strynadka, N.C, Withers, S.G, McIntosh, L.P. | | Deposit date: | 2012-10-15 | | Release date: | 2013-05-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Strategies for modulating the pH-dependent activity of a family 11 glycoside hydrolase
Biochemistry, 52, 2013
|
|
2KXX
 
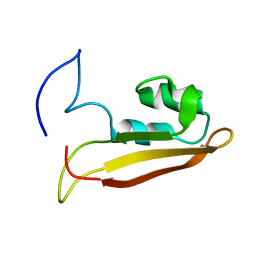 | | NMR Structure of Escherichia coli BamE, a Lipoprotein Component of the beta-Barrel Assembly Machinery Complex | | Descriptor: | Small protein A | | Authors: | Kim, K, Okon, M, Escobar, E, Kang, H, McIntosh, L, Paetzel, M. | | Deposit date: | 2010-05-13 | | Release date: | 2011-01-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of Escherichia coli BamE, a Lipoprotein Component of the beta-Barrel Assembly Machinery Complex.
Biochemistry, 50, 2011
|
|
2LQV
 
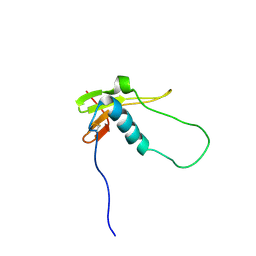 | | YebF | | Descriptor: | Protein yebF | | Authors: | Prehna, G, Zhang, G, Gong, X, Duszyk, M, Okon, M, Mcintosh, L.P, Weiner, J.H, Strynadka, N.C.J. | | Deposit date: | 2012-03-16 | | Release date: | 2012-06-13 | | Last modified: | 2024-11-27 | | Method: | SOLUTION NMR | | Cite: | A Protein Export Pathway Involving Escherichia coli Porins.
Structure, 20, 2012
|
|
2MD5
 
 | | Structure of uninhibited ETV6 ETS domain | | Descriptor: | Transcription factor ETV6 | | Authors: | De, S, Mcintosh, L.P, Chan, A.C, Coyne, H.J, Okon, M, Graves, B.J, Murphy, M.E. | | Deposit date: | 2013-08-29 | | Release date: | 2013-12-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Steric Mechanism of Auto-Inhibitory Regulation of Specific and Non-Specific DNA Binding by the ETS Transcriptional Repressor ETV6.
J.Mol.Biol., 426, 2014
|
|
2JW1
 
 | | Structural characterization of the type III pilotin-secretin interaction in Shigella flexneri by NMR spectroscopy | | Descriptor: | Lipoprotein mxiM, Outer membrane protein mxiD | | Authors: | Okon, M.S, Lario, P.I, Creagh, L, Jung, Y.M.T, Maurelli, A.T, Strynadka, N.C.J, McIntosh, L.P. | | Deposit date: | 2007-10-02 | | Release date: | 2008-09-02 | | Last modified: | 2024-11-27 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of the Type-III Pilot-Secretin Complex from Shigella flexneri
Structure, 16, 2008
|
|
3J6D
 
 | | Model of the PrgH-PrgK periplasmic rings | | Descriptor: | Pathogenicity 1 island effector protein, Protein PrgH | | Authors: | Bergeron, J.R.C, Strynadka, N.C.J. | | Deposit date: | 2014-02-14 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (11.7 Å) | | Cite: | The Modular Structure of the Inner-Membrane Ring Component PrgK Facilitates Assembly of the Type III Secretion System Basal Body.
Structure, 23, 2015
|
|
4W4M
 
 | | Crystal structure of PrgK 19-92 | | Descriptor: | Lipoprotein PrgK | | Authors: | Bergeron, J.R.C, Strynadka, N.C.J. | | Deposit date: | 2014-08-15 | | Release date: | 2014-10-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Modular Structure of the Inner-Membrane Ring Component PrgK Facilitates Assembly of the Type III Secretion System Basal Body.
Structure, 23, 2015
|
|
3LB9
 
 | | Crystal structure of the B. circulans cpA123 circular permutant | | Descriptor: | Endo-1,4-beta-xylanase | | Authors: | D'Angelo, I, Reitinger, S, Ludwiczek, M, Strynadka, N, Withers, S.G, Mcintosh, L.P. | | Deposit date: | 2010-01-08 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Circular permutation of Bacillus circulans xylanase: a kinetic and structural study.
Biochemistry, 49, 2010
|
|
6UM9
 
 | |
7JU2
 
 | | Crystal structure of the monomeric ETV6 PNT domain | | Descriptor: | FORMIC ACID, Transcription factor ETV6 | | Authors: | Gerak, C.A.N, Kolesnikov, M, Murphy, M.E.P, McIntosh, L.P. | | Deposit date: | 2020-08-19 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85002184 Å) | | Cite: | Biophysical characterization of the ETV6 PNT domain polymerization interfaces.
J.Biol.Chem., 296, 2021
|
|
