7F1I
 
 | | Designed enzyme RA61 M48K/I72D mutant: form II | | Descriptor: | Engineered Retroaldolase | | Authors: | Fujioka, T, Oka, M, Numoto, N, Ito, N, Oda, M, Tanaka, F. | | Deposit date: | 2021-06-09 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Varying the Directionality of Protein Catalysts for Aldol and Retro-Aldol Reactions.
Chembiochem, 23, 2022
|
|
7F1H
 
 | | Designed enzyme RA61 M48K/I72D mutant: form I | | Descriptor: | Engineered Retroaldolase, FORMIC ACID, GLYCEROL | | Authors: | Fujioka, T, Oka, M, Numoto, N, Ito, N, Oda, M, Tanaka, F. | | Deposit date: | 2021-06-09 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Varying the Directionality of Protein Catalysts for Aldol and Retro-Aldol Reactions.
Chembiochem, 23, 2022
|
|
7F1K
 
 | | Designed enzyme RA61 M48K/I72D mutant: form IV | | Descriptor: | Engineered Retroaldolase | | Authors: | Fujioka, T, Oka, M, Numoto, N, Ito, N, Oda, M, Tanaka, F. | | Deposit date: | 2021-06-09 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Varying the Directionality of Protein Catalysts for Aldol and Retro-Aldol Reactions.
Chembiochem, 23, 2022
|
|
7F1J
 
 | | Designed enzyme RA61 M48K/I72D mutant: form III | | Descriptor: | Engineered Retroaldolase | | Authors: | Fujioka, T, Oka, M, Numoto, N, Ito, N, Oda, M, Tanaka, F. | | Deposit date: | 2021-06-09 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Varying the Directionality of Protein Catalysts for Aldol and Retro-Aldol Reactions.
Chembiochem, 23, 2022
|
|
7F1L
 
 | | Designed enzyme RA61 M48K/I72D mutant: form V | | Descriptor: | CHLORIDE ION, Engineered Retroaldolase, IMIDAZOLE | | Authors: | Fujioka, T, Oka, M, Numoto, N, Ito, N, Oda, M, Tanaka, F. | | Deposit date: | 2021-06-09 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Varying the Directionality of Protein Catalysts for Aldol and Retro-Aldol Reactions.
Chembiochem, 23, 2022
|
|
7YRO
 
 | | Crystal structure of mango fucosyltransferase 13 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, Fucosyltransferase, ... | | Authors: | Okada, T, Teramoto, T, Ihara, H, Ikeda, Y, Kakuta, Y. | | Deposit date: | 2022-08-10 | | Release date: | 2023-08-16 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Crystal structure of mango alpha 1,3/ alpha 1,4-fucosyltransferase elucidates unique elements that regulate Lewis A-dominant oligosaccharide assembly.
Glycobiology, 34, 2024
|
|
5Z98
 
 | | Crystal Structure of the Primate APOBEC3H Dimer mediated by RNA Duplex | | Descriptor: | Apolipoprotein B mRNA editing enzyme catalytic polypeptide-like protein 3H, RNA (5'-R(*AP*UP*AP*CP*CP*CP*GP*GP*CP*A)-3'), RNA (5'-R(P*CP*UP*GP*CP*CP*GP*GP*GP*UP*A)-3'), ... | | Authors: | Matsuoka, T, Nagae, T, Ode, H, Watanabe, N, Iwatani, Y. | | Deposit date: | 2018-02-02 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of chimpanzee APOBEC3H dimerization stabilized by double-stranded RNA.
Nucleic Acids Res., 46, 2018
|
|
1WMN
 
 | | Crystal structure of topaquinone-containing amine oxidase activated by cobalt ion | | Descriptor: | COBALT (II) ION, Phenylethylamine oxidase | | Authors: | Okajima, T, Kishishita, S, Chiu, Y.C, Murakawa, T, Kim, M, Yamaguchi, H, Hirota, S, Kuroda, S, Tanizawa, K. | | Deposit date: | 2004-07-13 | | Release date: | 2005-08-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reinvestigation of metal ion specificity for quinone cofactor biogenesis in bacterial copper amine oxidase
Biochemistry, 44, 2005
|
|
1WMO
 
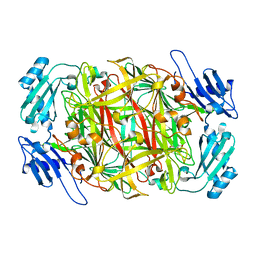 | | Crystal structure of topaquinone-containing amine oxidase activated by nickel ion | | Descriptor: | NICKEL (II) ION, Phenylethylamine oxidase | | Authors: | Okajima, T, Kishishita, S, Chiu, Y.C, Murakawa, T, Kim, M, Yamaguchi, H, Hirota, S, Kuroda, S, Tanizawa, K. | | Deposit date: | 2004-07-13 | | Release date: | 2005-08-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reinvestigation of metal ion specificity for quinone cofactor biogenesis in bacterial copper amine oxidase
Biochemistry, 44, 2005
|
|
1WMP
 
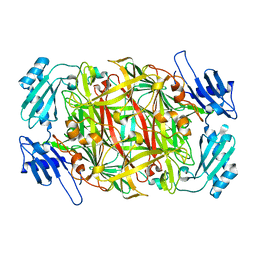 | | Crystal structure of amine oxidase complexed with cobalt ion | | Descriptor: | COBALT (II) ION, Phenylethylamine oxidase | | Authors: | Okajima, T, Kishishita, S, Chiu, Y.C, Murakawa, T, Kim, M, Yamaguchi, H, Hirota, S, Kuroda, S, Tanizawa, K. | | Deposit date: | 2004-07-13 | | Release date: | 2005-08-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reinvestigation of metal ion specificity for quinone cofactor biogenesis in bacterial copper amine oxidase
Biochemistry, 44, 2005
|
|
2D1V
 
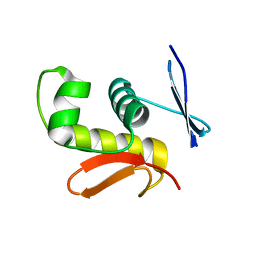 | | Crystal structure of DNA-binding domain of Bacillus subtilis YycF | | Descriptor: | Transcriptional regulatory protein yycF | | Authors: | Okajima, T, Okada, A, Watanabe, T, Yamamoto, K, Tanizawa, K, Utsumi, R. | | Deposit date: | 2005-09-01 | | Release date: | 2006-09-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Response regulator YycF essential for bacterial growth: X-ray crystal structure of the DNA-binding domain and its PhoB-like DNA recognition motif
Febs Lett., 582, 2008
|
|
7WEW
 
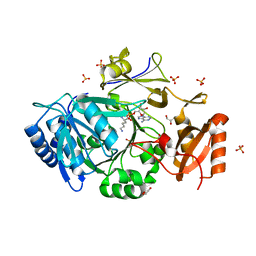 | | Structure of adenylation domain of epsilon-poly-L-lysine synthase | | Descriptor: | ADENOSINE-5'-[LYSYL-PHOSPHATE], Epsilon-poly-L-lysine synthase, GLYCEROL, ... | | Authors: | Okamoto, T, Yamanaka, K, Hamano, Y, Nagano, S, Hino, T. | | Deposit date: | 2021-12-24 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the adenylation domain from an epsilon-poly-l-lysine synthetase provides molecular mechanism for substrate specificity
Biochem.Biophys.Res.Commun., 596, 2022
|
|
1L9H
 
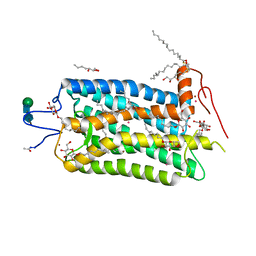 | | Crystal structure of bovine rhodopsin at 2.6 angstroms RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HEPTANE-1,2,3-TRIOL, MERCURY (II) ION, ... | | Authors: | Okada, T, Fujiyoshi, Y, Silow, M, Navarro, J, Landau, E.M, Shichida, Y. | | Deposit date: | 2002-03-23 | | Release date: | 2002-05-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Functional role of internal water molecules in rhodopsin revealed by X-ray crystallography.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2E0Y
 
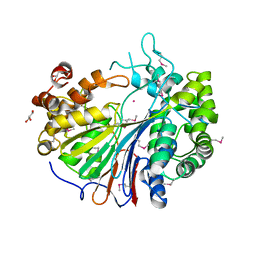 | |
6JI6
 
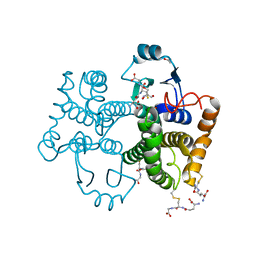 | |
2ZTJ
 
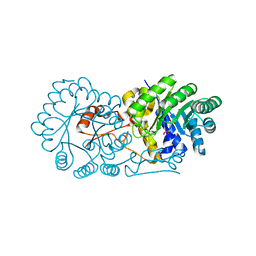 | | Crystal structure of homocitrate synthase from Thermus thermophilus complexed with alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, COPPER (II) ION, Homocitrate synthase | | Authors: | Okada, T, Tomita, T, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2008-10-06 | | Release date: | 2009-10-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of substrate recognition and insight into feedback inhibition of homocitrate synthase from Thermus thermophilus
J.Biol.Chem., 285, 2010
|
|
2ZYF
 
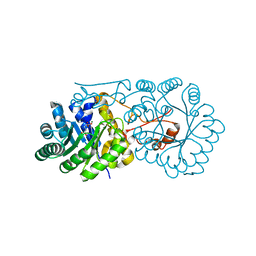 | | Crystal structure of homocitrate synthase from Thermus thermophilus complexed with magnesuim ion and alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, Homocitrate synthase, MAGNESIUM ION | | Authors: | Okada, T, Tomita, T, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2009-01-20 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Mechanism of substrate recognition and insight into feedback inhibition of homocitrate synthase from thermus thermophilus
J.Biol.Chem., 2009
|
|
3A9I
 
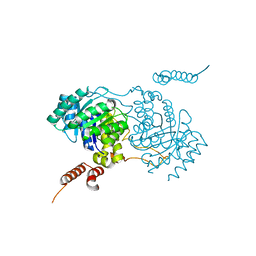 | | Crystal structure of homocitrate synthase from Thermus thermophilus complexed with Lys | | Descriptor: | COBALT (II) ION, Homocitrate synthase, LYSINE | | Authors: | Okada, T, Tomita, T, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2009-10-28 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of substrate recognition and insight into feedback inhibition of homocitrate synthase from Thermus thermophilus
J.Biol.Chem., 285, 2010
|
|
1U19
 
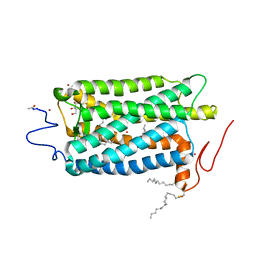 | | Crystal Structure of Bovine Rhodopsin at 2.2 Angstroms Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HEPTANE-1,2,3-TRIOL, MERCURY (II) ION, ... | | Authors: | Okada, T, Sugihara, M, Bondar, A.N, Elstner, M, Entel, P, Buss, V. | | Deposit date: | 2004-07-15 | | Release date: | 2004-10-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The retinal conformation and its environment in rhodopsin in light of a new 2.2 A crystal structure
J.Mol.Biol., 342, 2004
|
|
2ZTK
 
 | | Crystal structure of homocitrate synthase from Thermus thermophilus complexed with homocitrate | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, COPPER (II) ION, Homocitrate synthase | | Authors: | Okada, T, Tomita, T, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2008-10-06 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Mechanism of substrate recognition and insight into feedback inhibition of homocitrate synthase from Thermus thermophilus
J.Biol.Chem., 285, 2010
|
|
2E0X
 
 | |
2E0W
 
 | |
3AUU
 
 | | Crystal structure of Bacillus megaterium glucose dehydrogenase 4 in complex with D-glucose | | Descriptor: | Glucose 1-dehydrogenase 4, beta-D-glucopyranose | | Authors: | Nishioka, T, Yasutake, Y, Nishiya, Y, Tamura, T. | | Deposit date: | 2011-02-16 | | Release date: | 2012-02-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-guided mutagenesis for the improvement of substrate specificity of Bacillus megaterium glucose 1-dehydrogenase IV
Febs J., 279, 2012
|
|
2ZK7
 
 | | Structure of a C-terminal deletion mutant of Thermoplasma acidophilum aldohexose dehydrogenase (AldT) | | Descriptor: | Glucose 1-dehydrogenase related protein | | Authors: | Nishioka, T, Yasutake, Y, Nishiya, Y, Tamura, N, Tamura, T. | | Deposit date: | 2008-03-12 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | C-terminal tail derived from the neighboring subunit is critical for the activity of Thermoplasma acidophilum D-aldohexose dehydrogenase
Proteins, 74, 2009
|
|
3AUT
 
 | | Crystal structure of Bacillus megaterium glucose dehydrogenase 4 in complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Glucose 1-dehydrogenase 4 | | Authors: | Nishioka, T, Yasutake, Y, Nishiya, Y, Tamura, T. | | Deposit date: | 2011-02-16 | | Release date: | 2012-02-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-guided mutagenesis for the improvement of substrate specificity of Bacillus megaterium glucose 1-dehydrogenase IV
Febs J., 279, 2012
|
|
