1LDF
 
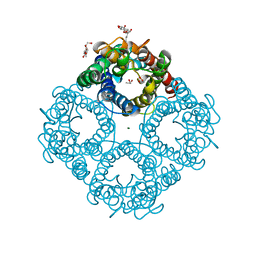 | | CRYSTAL STRUCTURE OF THE E. COLI GLYCEROL FACILITATOR (GLPF) MUTATION W48F, F200T | | Descriptor: | GLYCEROL, Glycerol uptake facilitator protein, MAGNESIUM ION, ... | | Authors: | Nollert, P, Miercke, L.J.W, O'Connell, J, Stroud, R.M. | | Deposit date: | 2002-04-08 | | Release date: | 2002-05-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Control of the selectivity of the aquaporin water channel family by global orientational tuning.
Science, 296, 2002
|
|
1LDA
 
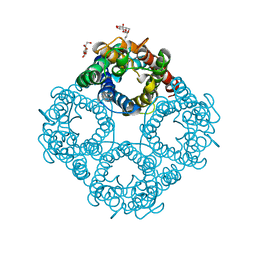 | | CRYSTAL STRUCTURE OF THE E. COLI GLYCEROL FACILITATOR (GLPF) WITHOUT SUBSTRATE GLYCEROL | | Descriptor: | Glycerol uptake facilitator protein, octyl beta-D-glucopyranoside | | Authors: | Nollert, P, Miercke, L.J.W, O'Connell, J, Stroud, R.M. | | Deposit date: | 2002-04-08 | | Release date: | 2002-05-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Control of the selectivity of the aquaporin water channel family by global orientational tuning.
Science, 296, 2002
|
|
1LDI
 
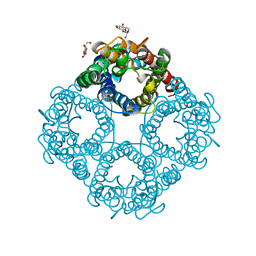 | | CRYSTAL STRUCTURE OF THE E. COLI GLYCEROL FACILITATOR (GLPF) WITHOUT SUBSTRATE GLYCEROL | | Descriptor: | Glycerol uptake facilitator protein, octyl beta-D-glucopyranoside | | Authors: | Nollert, P, Miercke, L.J.W, O'Connell, J, Stroud, R.M. | | Deposit date: | 2002-04-08 | | Release date: | 2002-05-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Control of the selectivity of the aquaporin water channel family by global orientational tuning.
Science, 296, 2002
|
|
1QKO
 
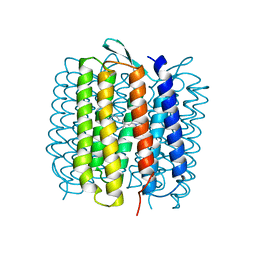 | | HIGH RESOLUTION X-RAY STRUCTURE OF AN EARLY INTERMEDIATE IN THE BACTERIORHODOPSIN PHOTOCYCLE | | Descriptor: | BACTERIORHODOPSIN, RETINAL | | Authors: | Edman, K, Nollert, P, Royant, A, Belrhali, H, Pebay-Peyroula, E, Hajdu, J, Neutze, R, Landau, E.M. | | Deposit date: | 1999-07-30 | | Release date: | 1999-10-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High Resolution X-Ray Structure of an Early Intermediate in the Bacteriorhodopsin Photocycle
Nature, 401, 1999
|
|
1QKP
 
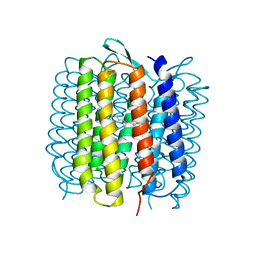 | | HIGH RESOLUTION X-RAY STRUCTURE OF AN EARLY INTERMEDIATE IN THE BACTERIORHODOPSIN PHOTOCYCLE | | Descriptor: | BACTERIORHODOPSIN, RETINAL | | Authors: | Edman, K, Nollert, P, Royant, A, Belrhali, H, Pebay-Peyroula, E, Hajdu, J, Neutze, R, Landau, E.M. | | Deposit date: | 1999-07-30 | | Release date: | 1999-10-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-resolution X-ray structure of an early intermediate in the bacteriorhodopsin photocycle.
Nature, 401, 1999
|
|
1GU8
 
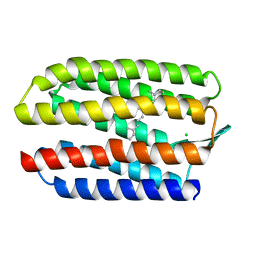 | | SENSORY RHODOPSIN II | | Descriptor: | CHLORIDE ION, RETINAL, SENSORY RHODOPSIN II | | Authors: | Edman, K, Royant, A, Nollert, P, Maxwell, C.A, Pebay-Peyroula, E, Navarro, J, Neutze, R, Landau, E.M. | | Deposit date: | 2002-01-24 | | Release date: | 2002-04-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Early Structural Rearrangements in the Photocycle of an Integral Membrane Sensory Receptor
Structure, 10, 2002
|
|
1H68
 
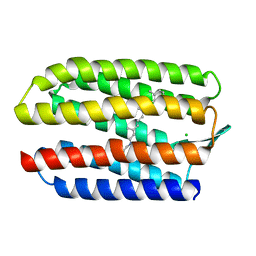 | | sensory rhodopsin II | | Descriptor: | CHLORIDE ION, RETINAL, SENSORY RHODOPSIN II | | Authors: | Royant, A, Nollert, P, Edman, K, Neutze, R, Landau, E.M, Pebay-Peyroula, E, Navarro, J. | | Deposit date: | 2001-06-08 | | Release date: | 2001-08-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-Ray Structure of Sensory Rhodopsin II at 2.1 A Resolution
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1QHJ
 
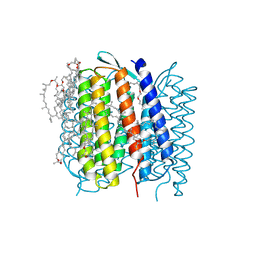 | | X-RAY STRUCTURE OF BACTERIORHODOPSIN GROWN IN LIPIDIC CUBIC PHASES | | Descriptor: | 1,2-[DI-2,6,10,14-TETRAMETHYL-HEXADECAN-16-OXY]-PROPANE, PROTEIN (BACTERIORHODOPSIN), RETINAL | | Authors: | Belrhali, H, Nollert, P, Royant, A, Menzel, C, Rosenbusch, J.P, Landau, E.M, Pebay-Peyroula, E. | | Deposit date: | 1999-05-04 | | Release date: | 1999-07-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein, lipid and water organization in bacteriorhodopsin crystals: a molecular view of the purple membrane at 1.9 A resolution.
Structure Fold.Des., 7, 1999
|
|
1FX8
 
 | | CRYSTAL STRUCTURE OF THE E. COLI GLYCEROL FACILITATOR (GLPF) WITH SUBSTRATE GLYCEROL | | Descriptor: | GLYCEROL, GLYCEROL UPTAKE FACILITATOR PROTEIN, octyl beta-D-glucopyranoside | | Authors: | Fu, D, Libson, A, Miercke, L.J.W, Weitzman, C, Nollert, P, Stroud, R.M. | | Deposit date: | 2000-09-25 | | Release date: | 2000-11-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a glycerol-conducting channel and the basis for its selectivity.
Science, 290, 2000
|
|
1GUE
 
 | | SENSORY RHODOPSIN II | | Descriptor: | CHLORIDE ION, RETINAL, SENSORY RHODOPSIN II | | Authors: | Edman, K, Royant, A, Nollert, P, Maxwell, C.A, Pebay-Peyroula, E, Navarro, J, Neutze, R, Landau, E.M. | | Deposit date: | 2002-01-24 | | Release date: | 2002-04-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Early Structural Rearrangements in the Photocycle of an Integral Membrane Sensory Receptor
Structure, 10, 2002
|
|
3R2V
 
 | |
3CXK
 
 | | 1.7 A Crystal structure of methionine-R-sulfoxide reductase from Burkholderia pseudomallei: crystallization in a microfluidic crystal card. | | Descriptor: | ACETATE ION, Methionine-R-sulfoxide reductase, ZINC ION | | Authors: | Lovell, S, Gerdts, C, Staker, B, Craigen, D, Stewart, L, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2008-04-24 | | Release date: | 2008-05-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The plug-based nanovolume Microcapillary Protein Crystallization System (MPCS).
Acta Crystallogr.,Sect.D, 64, 2008
|
|
