1G0D
 
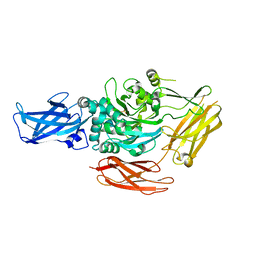 | | CRYSTAL STRUCTURE OF RED SEA BREAM TRANSGLUTAMINASE | | Descriptor: | PROTEIN-GLUTAMINE GAMMA-GLUTAMYLTRANSFERASE, SULFATE ION | | Authors: | Noguchi, K, Ishikawa, K, Yokoyama, K, Ohtsuka, T, Nio, N, Suzuki, E. | | Deposit date: | 2000-10-06 | | Release date: | 2001-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of red sea bream transglutaminase.
J.Biol.Chem., 276, 2001
|
|
7CG3
 
 | | Staggered ring conformation of CtHsp104 (Hsp104 from Chaetomium Thermophilum) | | Descriptor: | Heat shock protein 104 | | Authors: | Inoue, Y, Hanazono, Y, Noi, K, Kawamoto, A, Kimatsuka, M, Harada, R, Takeda, K, Iwamasa, N, Shibata, K, Noguchi, K, Shigeta, Y, Namba, K, Ogura, T, Miki, K, Shinohara, K, Yohda, M. | | Deposit date: | 2020-06-30 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Split conformation of Chaetomium thermophilum Hsp104 disaggregase.
Structure, 29, 2021
|
|
8IMR
 
 | | Structure of ligand-free human macrophage migration inhibitory factor | | Descriptor: | 1,2-ETHANEDIOL, ISOPROPYL ALCOHOL, Macrophage migration inhibitory factor, ... | | Authors: | Sugishima, K, Noguchi, K, Yohda, M, Odaka, M, Matsumura, H. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Identification of methotrexate as an inhibitor of macrophage migration inhibitory factor by high-resolution crystal structure analysis
To Be Published
|
|
3CI7
 
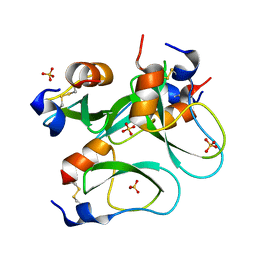 | | Crystal structure of a simplified BPTI containing 20 alanines | | Descriptor: | SULFATE ION, bovine pancreatic trypsin inhibitor | | Authors: | Islam, M.M, Sohya, S, Noguchi, K, Yohda, M, Kuroda, Y. | | Deposit date: | 2008-03-11 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of an extensively simplified variant of bovine pancreatic trypsin inhibitor in which over one-third of the residues are alanines
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1X1K
 
 | | Host-guest peptide (Pro-Pro-Gly)4-(Pro-alloHyp-Gly)-(Pro-Pro-Gly)4 | | Descriptor: | Host-guest peptide (Pro-Pro-Gly)4-(Pro-alloHyp-Gly)-(Pro-Pro-Gly)4 | | Authors: | Jiravanichanun, N, Hongo, C, Wu, G, Noguchi, K, Okuyama, K, Nishino, N, Silva, T. | | Deposit date: | 2005-04-05 | | Release date: | 2005-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Unexpected puckering of hydroxyproline in the guest triplets, hyp-pro-gly and pro-allohyp-gly sandwiched between pro-pro-gly sequence
Chembiochem, 6, 2005
|
|
7XTX
 
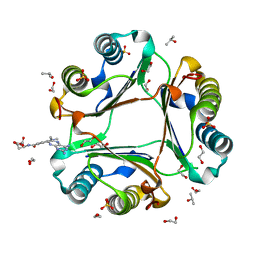 | | High resolution crystal structure of human macrophage migration inhibitory factor in complex with methotrexate | | Descriptor: | 1,2-ETHANEDIOL, ISOPROPYL ALCOHOL, Macrophage migration inhibitory factor, ... | | Authors: | Sugishima, K, Noguchi, K, Yohda, M, Odaka, M, Matsumura, H. | | Deposit date: | 2022-05-18 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Identification of methotrexate as an inhibitor of macrophage migration inhibitory factor by high-resolution crystal structure analysis
To Be Published
|
|
5ZUI
 
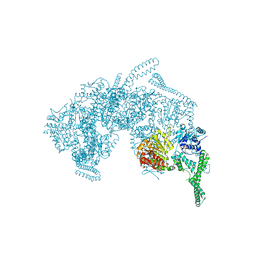 | | Crystal Structure of HSP104 from Chaetomium thermophilum | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Heat Shock Protein 104, SULFATE ION | | Authors: | Hanazono, Y, Inoue, Y, Noguchi, K, Yohda, M, Shinohara, K, Takeda, K, Miki, K. | | Deposit date: | 2018-05-07 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Split conformation of Chaetomium thermophilum Hsp104 disaggregase.
Structure, 2021
|
|
1V6Q
 
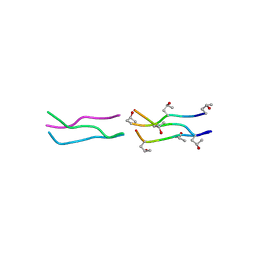 | | Crystal Structures of Collagen Model Peptides with Pro-Hyp-Gly Sequence at 1.3 A | | Descriptor: | Collagen like peptide | | Authors: | Okuyama, K, Hongo, C, Fukushima, R, Wu, G, Narita, H, Noguchi, K, Tanaka, Y, Nishino, N. | | Deposit date: | 2003-12-03 | | Release date: | 2004-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structures of collagen model peptides with Pro-Hyp-Gly repeating sequence at 1.26 A resolution: implications for proline ring puckering
Biopolymers, 76, 2004
|
|
1V4F
 
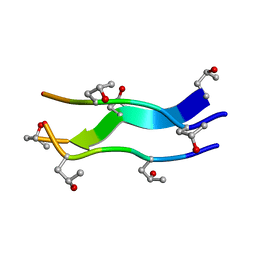 | | Crystal structures of collagen model peptides with pro-hyp-gly sequence at 1.3A | | Descriptor: | collagen like peptide | | Authors: | Okuyama, K, Hongo, C, Fukushima, R, Wu, G, Noguchi, K, Tanaka, Y, Nishino, N. | | Deposit date: | 2003-11-13 | | Release date: | 2004-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystal structures of collagen model peptides with Pro-Hyp-Gly repeating sequence at 1.26 A resolution: implications for proline ring puckering
Biopolymers, 76, 2004
|
|
1V7H
 
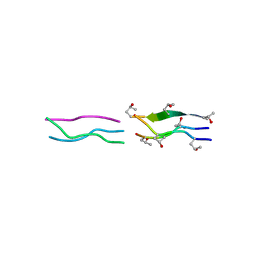 | | Crystal Structures of Collagen Model Peptides with Pro-Hyp-Gly Sequence at 1.26 A | | Descriptor: | Collagen like peptide | | Authors: | Okuyama, K, Hongo, C, Fukushima, R, Wu, G, Narita, H, Noguchi, K, Tanaka, Y, Nishino, N. | | Deposit date: | 2003-12-17 | | Release date: | 2004-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structures of collagen model peptides with Pro-Hyp-Gly repeating sequence at 1.26 A resolution: implications for proline ring puckering
Biopolymers, 76, 2004
|
|
1ITT
 
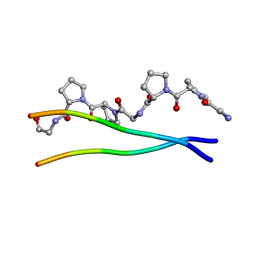 | | Average Crystal Structure of (Pro-Pro-Gly)9 at 1.0 angstroms Resolution | | Descriptor: | COLLAGEN TRIPLE HELIX | | Authors: | Hongo, C, Nagarajan, V, Noguchi, K, Kamitori, S, Okuyama, K, Tanaka, Y, Nishino, N. | | Deposit date: | 2002-02-03 | | Release date: | 2003-02-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Average Crystal Structure of (Pro-Pro-Gly)9 at 1.0 angstroms Resolution
Plym.J., 33, 2001
|
|
3WVE
 
 | | Crystal structure of Nitrile Hydratase mutant bR56K complexed with Trimethylacetonitrile, before photo-activation | | Descriptor: | 2,2-dimethylpropanenitrile, CHLORIDE ION, FE (III) ION, ... | | Authors: | Yamanaka, Y, Hashimoto, K, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2014-05-17 | | Release date: | 2015-06-17 | | Last modified: | 2015-11-04 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Time-Resolved Crystallography of the Reaction Intermediate of Nitrile Hydratase: Revealing a Role for the Cysteinesulfenic Acid Ligand as a Catalytic Nucleophile.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
3WVD
 
 | | Crystal structure of Nitrile Hydratase mutant bR56K complexed with Trimethylacetonitrile, photo-activated for 50 min | | Descriptor: | 2,2-dimethylpropanenitrile, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Yamanaka, Y, Hashimoto, K, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2014-05-17 | | Release date: | 2015-06-17 | | Last modified: | 2015-11-04 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Time-Resolved Crystallography of the Reaction Intermediate of Nitrile Hydratase: Revealing a Role for the Cysteinesulfenic Acid Ligand as a Catalytic Nucleophile.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
3X25
 
 | | Crystal structure of Nitrile Hydratase mutant bR56K complexed with Trimethylacetonitrile, photo-activated for 700 min | | Descriptor: | 2,2-dimethylpropanenitrile, CHLORIDE ION, FE (III) ION, ... | | Authors: | Yamanaka, Y, Hashimoto, K, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2014-12-10 | | Release date: | 2016-01-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Time-Resolved Crystallography of the Reaction Intermediate of Nitrile Hydratase: Revealing a Role for the Cysteinesulfenic Acid Ligand as a Catalytic Nucleophile.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
3X26
 
 | | Crystal structure of Nitrile Hydratase mutant bR56K complexed with Trimethylacetonitrile, photo-activated for 5 min | | Descriptor: | 2,2-dimethylpropanenitrile, CHLORIDE ION, FE (III) ION, ... | | Authors: | Yamanaka, Y, Hashimoto, K, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2014-12-10 | | Release date: | 2016-01-27 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Time-Resolved Crystallography of the Reaction Intermediate of Nitrile Hydratase: Revealing a Role for the Cysteinesulfenic Acid Ligand as a Catalytic Nucleophile.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
3X24
 
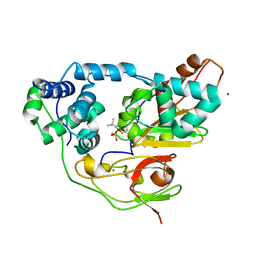 | | Crystal structure of Nitrile Hydratase mutant bR56K complexed with Trimethylacetonitrile, photo-activated for 120 min | | Descriptor: | 2,2-dimethylpropanenitrile, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Yamanaka, Y, Hashimoto, K, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2014-12-10 | | Release date: | 2016-01-27 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Time-Resolved Crystallography of the Reaction Intermediate of Nitrile Hydratase: Revealing a Role for the Cysteinesulfenic Acid Ligand as a Catalytic Nucleophile.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6L98
 
 | | Crystalline cast nephropathy-causing Bence-Jones protein AK: An entire immunoglobulin lambda light chain dimer | | Descriptor: | Bence-Jones protein lambda light chain AK | | Authors: | Nakagaki, T, Noguchi, K, Yohda, M, Odaka, M, Wakui, H, Matsumura, H. | | Deposit date: | 2019-11-08 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Multiple Myeloma-Associated Ig Light Chain Crystalline Cast Nephropathy.
Kidney Int Rep, 5, 2020
|
|
2D3H
 
 | | Crystal structures of collagen model peptides (Pro-Pro-Gly)4-Hyp-Hyp-Gly-(Pro-Pro-Gly)4 | | Descriptor: | COLLAGEN MODEL PEPTIDES (PRO-PRO-GLY)4-HYP-HYP-GLY-(PRO-PRO-GLY)4 | | Authors: | Wu, G, Noguchi, K, Okuyama, K, Mizuno, K, Bachinger, H.P. | | Deposit date: | 2005-09-28 | | Release date: | 2006-09-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | High-resolution structures of collagen-like peptides [(Pro-Pro-Gly)(4)-Xaa-Yaa-Gly-(Pro-Pro-Gly)(4)]: Implications for triple-helix hydration and Hyp(X) puckering.
Biopolymers, 91, 2009
|
|
2D3F
 
 | | Crystal structures of collagen model peptides (Pro-Pro-Gly)4-Pro-Hyp-Gly-(Pro-Pro-Gly)4 | | Descriptor: | COLLAGEN MODEL PEPTIDES (PRO-PRO-GLY)4-PRO-HYP-GLY-(PRO-PRO-GLY)4 | | Authors: | Wu, G, Noguchi, K, Okuyama, K, Ebisuzaki, S, Nishino, N. | | Deposit date: | 2005-09-27 | | Release date: | 2006-09-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | High-resolution structures of collagen-like peptides [(Pro-Pro-Gly)(4)-Xaa-Yaa-Gly-(Pro-Pro-Gly)(4)]: Implications for triple-helix hydration and Hyp(X) puckering.
Biopolymers, 91, 2009
|
|
4Z9F
 
 | | Halohydrin hydrogen-halide-lyase, HheA | | Descriptor: | CHLORIDE ION, Halohydrin epoxidase A | | Authors: | Watanabe, F, Yu, F, Ohtaki, A, Yamanaka, Y, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2015-04-10 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of halohydrin hydrogen-halide-lyases from Corynebacterium sp. N-1074
Proteins, 83, 2015
|
|
4ZU3
 
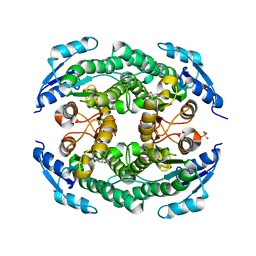 | | Halohydrin hydrogen-halide-lyases, HheB | | Descriptor: | 3-hydroxypentanedinitrile, Halohydrin epoxidase B, MAGNESIUM ION, ... | | Authors: | Watanabe, F, Yu, F, Ohtaki, A, Yamanaka, Y, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2015-05-15 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of halohydrin hydrogen-halide-lyases from Corynebacterium sp. N-1074
Proteins, 83, 2015
|
|
4ZD6
 
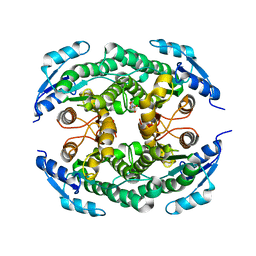 | | Halohydrin hydrogen-halide-lyase, HheB | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Halohydrin epoxidase B, ... | | Authors: | Watanabe, F, Yu, F, Ohtaki, A, Yamanaka, Y, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2015-04-17 | | Release date: | 2015-11-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of halohydrin hydrogen-halide-lyases from Corynebacterium sp. N-1074
Proteins, 83, 2015
|
|
2CUO
 
 | | Collagen model peptide (PRO-PRO-GLY)9 | | Descriptor: | COLLAGEN MODEL PEPTIDE (PRO-PRO-GLY)9 | | Authors: | Hongo, C, Noguchi, K, Okuyama, K, Tanaka, Y, Nishino, N. | | Deposit date: | 2005-05-27 | | Release date: | 2005-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Repetitive interactions observed in the crystal structure of a collagen-model peptide, [(Pro-Pro-Gly)9]3
J.Biochem.(Tokyo), 138, 2005
|
|
5B19
 
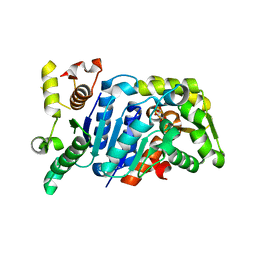 | | Picrophilus torridus aspartate racemase | | Descriptor: | Aspartate racemase, L(+)-TARTARIC ACID | | Authors: | Aihara, T, Ito, T, Yamanaka, Y, Noguchi, K, Odaka, M, Sekine, M, Homma, H, Yohda, M. | | Deposit date: | 2015-11-30 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Structural and functional characterization of aspartate racemase from the acidothermophilic archaeon Picrophilus torridus
Extremophiles, 20, 2016
|
|
5B12
 
 | | Crystal structure of the B-type halohydrin hydrogen-halide-lyase mutant F71W/Q125T/D199H from Corynebacterium sp. N-1074 | | Descriptor: | CHLORIDE ION, Halohydrin epoxidase B | | Authors: | Watanabe, F, Yu, F, Ohtaki, A, Yamanaka, Y, Noguchi, K, Odaka, M, Yohda, M. | | Deposit date: | 2015-11-17 | | Release date: | 2016-08-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.721 Å) | | Cite: | Improvement of enantioselectivity of the B-type halohydrin hydrogen-halide-lyase from Corynebacterium sp. N-1074
J.Biosci.Bioeng., 122, 2016
|
|
