2AI5
 
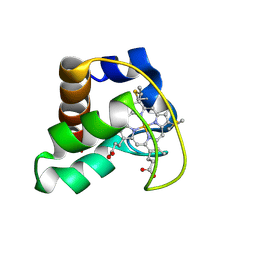 | | Solution Structure of Cytochrome C552, determined by Distributed Computing Implementation for NMR data | | Descriptor: | Cytochrome c-552, HEME C | | Authors: | Nakamura, S, Ichiki, S.I, Takashima, H, Uchiyama, S, Hasegawa, J, Kobayashi, Y, Sambongi, Y, Ohkubo, T. | | Deposit date: | 2005-07-29 | | Release date: | 2006-05-23 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structure of Cytochrome c552 from a Moderate Thermophilic Bacterium, Hydrogenophilus thermoluteolus: Comparative Study on the Thermostability of Cytochrome c
Biochemistry, 45, 2006
|
|
8IUA
 
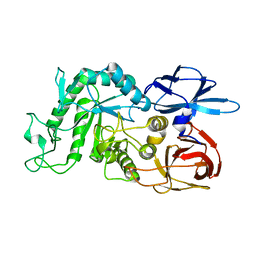 | |
8IU9
 
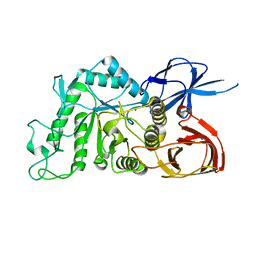 | |
8IU8
 
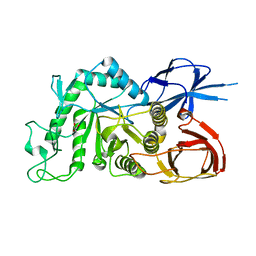 | |
8IUB
 
 | |
8IUC
 
 | |
6AKG
 
 | |
6AKE
 
 | |
6AKF
 
 | |
2DKN
 
 | |
2D0S
 
 | | Crystal structure of the Cytochrome C552 from moderate thermophilic bacterium, hydrogenophilus thermoluteolus | | Descriptor: | HEME C, cytochrome c | | Authors: | Nakamura, S, Ichiki, S.I, Takashima, H, Uchiyama, S, Hasegawa, J, Kobayashi, Y, Sambongi, Y, Ohkubo, T. | | Deposit date: | 2005-08-08 | | Release date: | 2006-05-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Cytochrome c552 from a Moderate Thermophilic Bacterium, Hydrogenophilus thermoluteolus: Comparative Study on the Thermostability of Cytochrome c
Biochemistry, 45, 2006
|
|
7FE3
 
 | |
7FE4
 
 | |
8WG1
 
 | | Crystal structure of GH97 glucodextranase mutant E509Q from Flavobacterium johnsoniae in complex with panose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Candidate alpha-glucosidase Glycoside hydrolase family 97, ... | | Authors: | Kurata, R, Nakamura, S, Miyazaki, T. | | Deposit date: | 2023-09-20 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural insights into alpha-(1→6)-linkage preference of GH97 glucodextranase from Flavobacterium johnsoniae.
Febs J., 291, 2024
|
|
8WG0
 
 | | Crystal structure of GH97 glucodextranase from Flavobacterium johnsoniae in complex with glucose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Candidate alpha-glucosidase Glycoside hydrolase family 97, ... | | Authors: | Kurata, R, Nakamura, S, Miyazaki, T. | | Deposit date: | 2023-09-20 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insights into alpha-(1→6)-linkage preference of GH97 glucodextranase from Flavobacterium johnsoniae.
Febs J., 291, 2024
|
|
8WG2
 
 | | Crystal structure of GH97 glucodextranase mutant E509Q from Flavobacterium johnsoniae in complex with isomaltotriose | | Descriptor: | CALCIUM ION, Candidate alpha-glucosidase Glycoside hydrolase family 97, alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose, ... | | Authors: | Kurata, R, Nakamura, S, Miyazaki, T. | | Deposit date: | 2023-09-20 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural insights into alpha-(1→6)-linkage preference of GH97 glucodextranase from Flavobacterium johnsoniae.
Febs J., 291, 2024
|
|
7C95
 
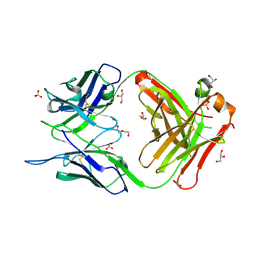 | | Crystal structure of the anti-human podoplanin antibody Fab fragment | | Descriptor: | GLYCEROL, Heavy chain of Fab fragment, Light chain of Fab fragment, ... | | Authors: | Nakamura, S, Suzuki, K, Ogasawara, S, Naruchi, K, Shimabukuro, J, Tukahara, N, Kaneko, M.K, Kato, Y, Murata, T. | | Deposit date: | 2020-06-04 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structure of an anti-podoplanin antibody bound to a disialylated O-linked glycopeptide.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
2Z2T
 
 | |
5XL0
 
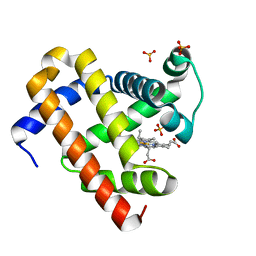 | | met-aquo form of sperm whale myoglobin reconstituted with 7-PF, a heme possesseing CF3 group as side chain | | Descriptor: | Myoglobin, SULFATE ION, fluorinated heme | | Authors: | Kanai, Y, Harada, A, Shibata, T, Nishimura, R, Namiki, K, Watanabe, M, Nakamura, S, Yumoto, F, Senda, T, Suzuki, A, Neya, S, Yamamoto, Y. | | Deposit date: | 2017-05-10 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Characterization of Heme Orientational Disorder in a Myoglobin Reconstituted with a Trifluoromethyl-Group-Substituted Heme Cofactor
Biochemistry, 56, 2017
|
|
2HYK
 
 | | The crystal structure of an endo-beta-1,3-glucanase from alkaliphilic Nocardiopsis sp.strain F96 | | Descriptor: | Beta-1,3-glucanase, CALCIUM ION, ETHANOL, ... | | Authors: | Fibriansah, G, Nakamura, S, Kumasaka, T. | | Deposit date: | 2006-08-07 | | Release date: | 2007-10-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The 1.3 A crystal structure of a novel endo-beta-1,3-glucanase of glycoside hydrolase family 16 from alkaliphilic Nocardiopsis sp. strain F96.
Proteins, 69, 2007
|
|
8Y1M
 
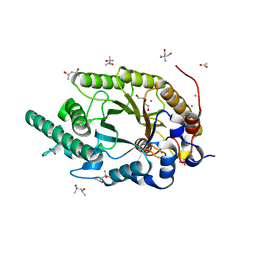 | | Xylanase R from Bacillus sp. TAR-1 complexed with xylobiose. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Nakamura, T, Kuwata, K, Takita, T, Mizutani, K, Mikami, B, Nakamura, S, Yasukawa, K. | | Deposit date: | 2024-01-25 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Activity-stability trade-off observed in variants at position 315 of the GH10 xylanase XynR.
Sci Rep, 14, 2024
|
|
8XY0
 
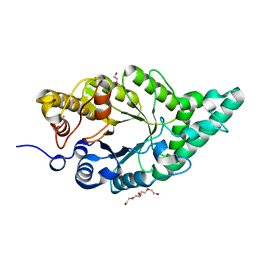 | | Activity-stability trade-off observed in variants at position 315 of the GH10 xylanase XynR | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Endo-1,4-beta-xylanase A | | Authors: | Nakamura, T, Takita, T, Mizutani, K, Mikami, B, Nakamura, S, Yasukawa, K. | | Deposit date: | 2024-01-19 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Activity-stability trade-off observed in variants at position 315 of the GH10 xylanase XynR.
Sci Rep, 14, 2024
|
|
4U4P
 
 | | Crystal structure of the human condensin SMC hinge domain heterodimer with short coiled coils | | Descriptor: | Structural maintenance of chromosomes protein 2, Structural maintenance of chromosomes protein 4 | | Authors: | Uchiyama, S, Kawahara, K, Hosokawa, Y, Fukakusa, S, Oki, H, Nakamura, S, Noda, M, Takino, R, Miyahara, Y, Maruno, T, Kobayashi, Y, Ohkubo, T, Fukui, K. | | Deposit date: | 2014-07-24 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis for dimer information and DNA recognition of human SMC proteins
to be published
|
|
5H03
 
 | | Crystal structure of an ADP-ribosylating toxin BECa from C. perfringens | | Descriptor: | Binary enterotoxin of Clostridium perfringens component a | | Authors: | Kawahara, K, Yonogi, S, Munetomo, R, Oki, H, Yoshida, T, Ohkubo, T, Kumeda, Y, Matsuda, S, Kodama, T, Iida, T, Nakamura, S. | | Deposit date: | 2016-10-03 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of the ADP-ribosylating component of BEC, the binary enterotoxin of Clostridium perfringens.
Biochem.Biophys.Res.Commun., 480, 2016
|
|
5H04
 
 | | Crystal structure of an ADP-ribosylating toxin BECa of a novel binary enterotoxin of C. perfringens with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Binary enterotoxin of Clostridium perfringens component a | | Authors: | Kawahara, K, Yonogi, S, Munetomo, R, Oki, H, Yoshida, T, Ohkubo, T, Kumeda, Y, Matsuda, S, Kodama, T, Iida, T, Nakamura, S. | | Deposit date: | 2016-10-03 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.825 Å) | | Cite: | Crystal structure of the ADP-ribosylating component of BEC, the binary enterotoxin of Clostridium perfringens.
Biochem.Biophys.Res.Commun., 480, 2016
|
|
