2IUF
 
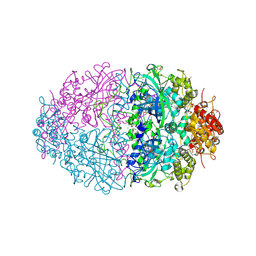 | | The structures of Penicillium vitale catalase: resting state, oxidised state (compound I) and complex with aminotriazole | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Murshudov, G, Borovik, A, Grebenko, A, Barynin, V, Vagin, A, Melik-Adamyan, W. | | Deposit date: | 2006-06-02 | | Release date: | 2006-07-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | The Structures and Electronic Configuration of Compound I Intermediates of Helicobacter Pylori and Penicillium Vitale Catalases Determined by X-Ray Crystallography and Qm/Mm Density Functional Theory Calculations.
J.Am.Chem.Soc., 129, 2007
|
|
3J7Y
 
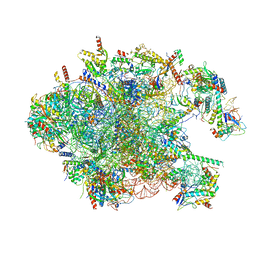 | | Structure of the large ribosomal subunit from human mitochondria | | Descriptor: | 16S rRNA, ADENOSINE MONOPHOSPHATE, CRIF1, ... | | Authors: | Brown, A, Amunts, A, Bai, X.C, Sugimoto, Y, Edwards, P.C, Murshudov, G, Scheres, S.H.W, Ramakrishnan, V. | | Deposit date: | 2014-08-26 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the large ribosomal subunit from human mitochondria.
Science, 346, 2014
|
|
8CKA
 
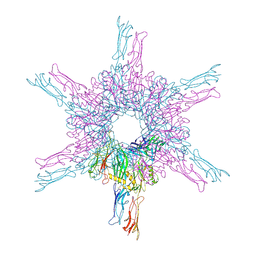 | | Deinococcus radidurans HPI S-layer | | Descriptor: | Hexagonally packed intermediate-layer surface protein | | Authors: | von Kuegelgen, A, Yamashita, K, Murshudov, G, Bharat, T. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Interdigitated immunoglobulin arrays form the hyperstable surface layer of the extremophilic bacterium Deinococcus radiodurans.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7A4M
 
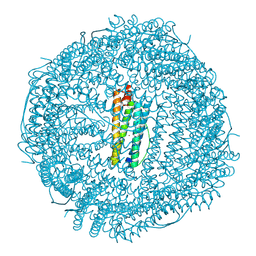 | | Cryo-EM structure of mouse heavy-chain apoferritin at 1.22 A | | Descriptor: | FE (III) ION, Ferritin heavy chain, ZINC ION | | Authors: | Nakane, T, Kotecha, A, Sente, A, Yamashita, K, McMullan, G, Masiulis, S, Brown, P.M.G.E, Grigoras, I.T, Malinauskaite, L, Malinauskas, T, Miehling, J, Yu, L, Karia, D, Pechnikova, E.V, de Jong, E, Keizer, J, Bischoff, M, McCormack, J, Tiemeijer, P, Hardwick, S.W, Chirgadze, D.Y, Murshudov, G, Aricescu, A.R, Scheres, S.H.W. | | Deposit date: | 2020-08-20 | | Release date: | 2020-10-28 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (1.22 Å) | | Cite: | Single-particle cryo-EM at atomic resolution.
Nature, 587, 2020
|
|
7A5V
 
 | | CryoEM structure of a human gamma-aminobutyric acid receptor, the GABA(A)R-beta3 homopentamer, in complex with histamine and megabody Mb25 in lipid nanodisc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Nakane, T, Kotecha, A, Sente, A, Yamashita, K, McMullan, G, Masiulis, S, Brown, P.M.G.E, Grigoras, I.T, Malinauskaite, L, Malinauskas, T, Miehling, J, Yu, L, Karia, D, Pechnikova, E.V, de Jong, E, Keizer, J, Bischoff, M, McCormack, J, Tiemeijer, P, Hardwick, S.W, Chirgadze, D.Y, Murshudov, G, Aricescu, A.R, Scheres, S.H.W. | | Deposit date: | 2020-08-22 | | Release date: | 2020-11-18 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (1.7 Å) | | Cite: | Single-particle cryo-EM at atomic resolution.
Nature, 587, 2020
|
|
6GX5
 
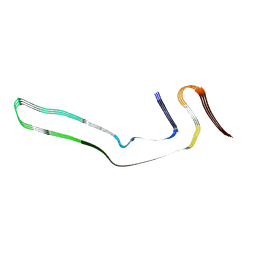 | | Narrow Pick Filament from Pick's disease brain | | Descriptor: | Microtubule-associated protein tau | | Authors: | Falcon, B, Zhang, W, Murzin, A.G, Murshudov, G, Garringer, H.J, Vidal, R, Crowther, R.A, Ghetti, B, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2018-06-26 | | Release date: | 2018-09-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of filaments from Pick's disease reveal a novel tau protein fold.
Nature, 561, 2018
|
|
1CZ1
 
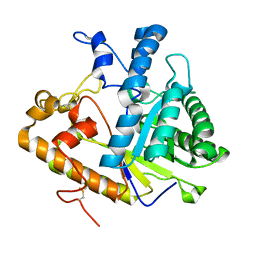 | | EXO-B-(1,3)-GLUCANASE FROM CANDIDA ALBICANS AT 1.85 A RESOLUTION | | Descriptor: | PROTEIN (EXO-B-(1,3)-GLUCANASE) | | Authors: | Cutfield, S.M, Davies, G.J, Murshudov, G, Anderson, B.F, Moody, P.C.E, Sullivan, P.A, Cutfield, J.F. | | Deposit date: | 1999-09-01 | | Release date: | 2000-01-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of the exo-beta-(1,3)-glucanase from Candida albicans in native and bound forms: relationship between a pocket and groove in family 5 glycosyl hydrolases.
J.Mol.Biol., 294, 1999
|
|
1EQC
 
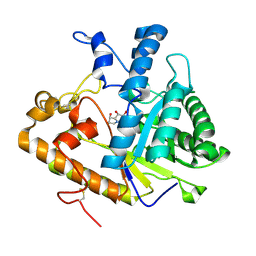 | | EXO-B-(1,3)-GLUCANASE FROM CANDIDA ALBICANS IN COMPLEX WITH CASTANOSPERMINE AT 1.85 A | | Descriptor: | CASTANOSPERMINE, EXO-(B)-(1,3)-GLUCANASE | | Authors: | Cutfield, S.M, Davies, G.J, Murshudov, G, Anderson, B.F, Moody, P.C.E, Sullivan, P.A, Cutfield, J.F. | | Deposit date: | 2000-04-03 | | Release date: | 2000-10-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of the exo-beta-(1,3)-glucanase from Candida albicans in native and bound forms: relationship between a pocket and groove in family 5 glycosyl hydrolases.
J.Mol.Biol., 294, 1999
|
|
1H02
 
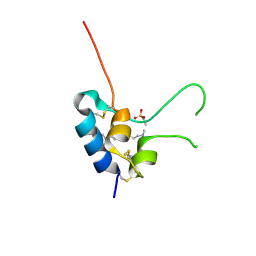 | | Human Insulin-like growth factor; SRS Daresbury data | | Descriptor: | INSULIN-LIKE GROWTH FACTOR I, N-DODECYL-N,N-DIMETHYL-3-AMMONIO-1-PROPANESULFONATE | | Authors: | Brzozowski, A.M, Dodson, E.J, Dodson, G.G, Murshudov, G, Verma, C, Turkenburg, J.P, De Bree, F.M, Dauter, Z. | | Deposit date: | 2002-06-11 | | Release date: | 2002-07-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Origins of the Functional Divergence of Human Insulin-Like Growth Factor-I and Insulin
Biochemistry, 41, 2002
|
|
1GZR
 
 | | Human Insulin-like growth factor; ESRF data | | Descriptor: | INSULIN-LIKE GROWTH FACTOR I, N-DODECYL-N,N-DIMETHYL-3-AMMONIO-1-PROPANESULFONATE | | Authors: | Brzozowski, A.M, Dodson, E.J, Dodson, G.G, Murshudov, G, Verma, C, Turkenburg, J.P, De Bree, F.M, Dauter, Z. | | Deposit date: | 2002-05-28 | | Release date: | 2002-07-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Origins of the Functional Divergence of Human Insulin-Like Growth Factor-I and Insulin
Biochemistry, 41, 2002
|
|
1GZZ
 
 | | Human Insulin-like growth factor; Hamburg data | | Descriptor: | INSULIN-LIKE GROWTH FACTOR I, N-DODECYL-N,N-DIMETHYL-3-AMMONIO-1-PROPANESULFONATE | | Authors: | Brzozowski, A.M, Dodson, E.J, Dodson, G.G, Murshudov, G, Verma, C, Turkenburg, J.P, De Bree, F.M, Dauter, Z. | | Deposit date: | 2002-06-10 | | Release date: | 2002-07-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Origins of the Functional Divergence of Human Insulin-Like Growth Factor-I and Insulin
Biochemistry, 41, 2002
|
|
1GZY
 
 | | Human Insulin-like growth factor; In-house data | | Descriptor: | INSULIN-LIKE GROWTH FACTOR I, N-DODECYL-N,N-DIMETHYL-3-AMMONIO-1-PROPANESULFONATE | | Authors: | Brzozowski, A.M, Dodson, E.J, Dodson, G.G, Murshudov, G, Verma, C, Turkenburg, J.P, De Bree, F.M, Dauter, Z. | | Deposit date: | 2002-06-10 | | Release date: | 2002-07-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural Origins of the Functional Divergence of Human Insulin-Like Growth Factor-I and Insulin
Biochemistry, 41, 2002
|
|
1QNO
 
 | | The 3-D structure of a Trichoderma reesei b-mannanase from glycoside hydrolase family 5 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDO-1,4-B-D-MANNANASE | | Authors: | Sabini, E, Schubert, H, Murshudov, G, Wilson, K.S, Siika-Aho, M, Penttila, M. | | Deposit date: | 1999-10-20 | | Release date: | 2000-10-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Three-Dimensional Structure of a Trichoderma Reesei Beta-Mannanase from Glycoside Hydrolase Family 5.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1QNP
 
 | | The 3-D structure of a Trichoderma reesei b-mannanase from glycoside hydrolase family 5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDO-1,4-B-D-MANNANASE, GLYCEROL, ... | | Authors: | Sabini, E, Schubert, H, Murshudov, G, Wilson, K.S, Siika-Aho, M, Penttila, M. | | Deposit date: | 1999-10-20 | | Release date: | 2000-10-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Three-Dimensional Structure of a Trichoderma Reesei Beta-Mannanase from Glycoside Hydrolase Family 5.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1QNR
 
 | | The 3-D structure of a Trichoderma reesei b-mannanase from glycoside hydrolase family 5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDO-1,4-B-D-MANNANASE, GLYCEROL, ... | | Authors: | Sabini, E, Schubert, H, Murshudov, G, Wilson, K.S, Siika-Aho, M, Penttila, M. | | Deposit date: | 1999-10-20 | | Release date: | 2000-10-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Three-Dimensional Structure of a Trichoderma Reesei Beta-Mannanase from Glycoside Hydrolase Family 5.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1QNQ
 
 | | The 3-D structure of a Trichoderma reesei b-mannanase from glycoside hydrolase family 5 | | Descriptor: | 2,2':6',2''-TERPYRIDINE PLATINUM(II) Chloride, 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDO-1,4-B-D-MANNANASE, ... | | Authors: | Sabini, E, Schubert, H, Murshudov, G, Wilson, K.S, Siika-Aho, M, Penttila, M. | | Deposit date: | 1999-10-20 | | Release date: | 2000-10-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Three-Dimensional Structure of a Trichoderma Reesei Beta-Mannanase from Glycoside Hydrolase Family 5.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1QNS
 
 | | The 3-D structure of a Trichoderma reesei b-mannanase from glycoside hydrolase family 5 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDO-1,4-B-D-MANNANASE, ... | | Authors: | Sabini, E, Schubert, H, Murshudov, G, Wilson, K.S, Siika-Aho, M, Penttila, M. | | Deposit date: | 1999-10-20 | | Release date: | 2000-10-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Three-Dimensional Structure of a Trichoderma Reesei Beta-Mannanase from Glycoside Hydrolase Family 5
Acta Crystallogr.,Sect.D, 56, 2000
|
|
4NE4
 
 | | Crystal structure of ABC transporter substrate binding protein ProX from Agrobacterium tumefaciens cocrystalized with BTB | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ABC transporter, substrate binding protein (Proline/glycine/betaine), ... | | Authors: | Tkaczuk, K.L, Nicholls, R, Kagan, O, Chruszcz, M, Domagalski, M.J, Savchenko, A, Joachimiak, A, Murshudov, G, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-10-28 | | Release date: | 2013-11-27 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of ABC transporter substrate binding protein ProX from Agrobacterium tumefaciens cocrystalized with BTB
To be Published
|
|
4ND9
 
 | | The putative substrate binding domain of ABC-type transporter from Agrobacterium tumefaciens in open conformation | | Descriptor: | ABC transporter, substrate binding protein (Proline/glycine/betaine) | | Authors: | Nicholls, R, Tkaczuk, K.L, Kagan, O, Chruszcz, M, Domagalski, M.J, Savchenko, A, Joachimiak, A, Murshudov, G, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-10-25 | | Release date: | 2013-12-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The putative substrate binding domain of ABC-type transporter from Agrobacterium tumefaciens in open conformation
To be Published
|
|
5O3O
 
 | | Pronase-treated paired helical filament in Alzheimer's disease brain | | Descriptor: | Microtubule-associated protein tau | | Authors: | Fitzpatrick, A.W.P, Falcon, B, He, S, Murzin, A.G, Murshudov, G, Garringer, H.G, Crowther, R.A, Ghetti, B, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2017-05-24 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of tau filaments from Alzheimer's disease.
Nature, 547, 2017
|
|
5O3L
 
 | | Paired helical filament in Alzheimer's disease brain | | Descriptor: | Microtubule-associated protein tau | | Authors: | Fitzpatrick, A.W.P, Falcon, B, He, S, Murzin, A.G, Murshudov, G, Garringer, H.G, Crowther, R.A, Ghetti, B, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2017-05-24 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of tau filaments from Alzheimer's disease.
Nature, 547, 2017
|
|
3J6B
 
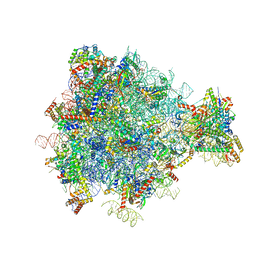 | | Structure of the yeast mitochondrial large ribosomal subunit | | Descriptor: | 21S ribosomal RNA, 54S ribosomal protein IMG1, mitochondrial, ... | | Authors: | Amunts, A, Brown, A, Bai, X.C, Llacer, J.L, Hussain, T, Emsley, P, Long, F, Murshudov, G, Scheres, S.H.W, Ramakrishnan, V. | | Deposit date: | 2014-01-22 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the yeast mitochondrial large ribosomal subunit.
Science, 343, 2014
|
|
5O3T
 
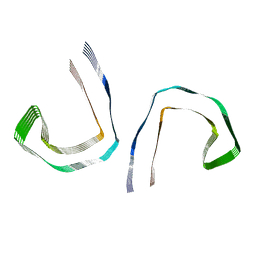 | | Straight filament in Alzheimer's disease brain | | Descriptor: | Microtubule-associated protein tau | | Authors: | Fitzpatrick, A.W.P, Falcon, B, He, S, Murzin, A.G, Murshudov, G, Garringer, H.G, Crowther, R.A, Ghetti, B, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2017-05-24 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of tau filaments from Alzheimer's disease.
Nature, 547, 2017
|
|
8RK1
 
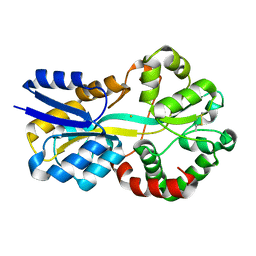 | | Crystal structure of FutA bound to Fe(III) solved by neutron diffraction | | Descriptor: | FE (III) ION, Putative iron ABC transporter, substrate binding protein | | Authors: | Bolton, R, Tews, I. | | Deposit date: | 2023-12-22 | | Release date: | 2024-01-17 | | Last modified: | 2024-10-16 | | Method: | NEUTRON DIFFRACTION (2.095 Å) | | Cite: | A redox switch allows binding of Fe(II) and Fe(III) ions in the cyanobacterial iron-binding protein FutA from Prochlorococcus.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8C4Y
 
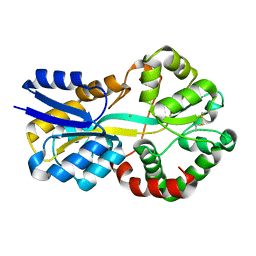 | | SFX structure of FutA bound to Fe(III) | | Descriptor: | FE (III) ION, Putative iron ABC transporter, substrate binding protein | | Authors: | Bolton, R, Tews, I. | | Deposit date: | 2023-01-05 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A redox switch allows binding of Fe(II) and Fe(III) ions in the cyanobacterial iron-binding protein FutA from Prochlorococcus.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
