7ZSG
 
 | | Structure of Orange Carotenoid Protein with canthaxanthin bound after 1 minute of illumination | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Chukhutsina, V.U, Baxter, J.M, Fadini, A, Morgan, R.M, Pope, M.A, Maghlaoui, K, Orr, C, Wagner, A, van Thor, J.J. | | Deposit date: | 2022-05-06 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Light activation of Orange Carotenoid Protein reveals bicycle-pedal single-bond isomerization.
Nat Commun, 13, 2022
|
|
7ZSJ
 
 | | Structure of Orange Carotenoid Protein with canthaxanthin bound after 10 minutes of illumination | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Chukhutsina, V.U, Baxter, J.M, Fadini, A, Morgan, R.M, Pope, M.A, Maghlaoui, K, Orr, C, Wagner, A, van Thor, J.J. | | Deposit date: | 2022-05-06 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Light activation of Orange Carotenoid Protein reveals bicycle-pedal single-bond isomerization.
Nat Commun, 13, 2022
|
|
7ZSF
 
 | | Structure of Orange Carotenoid Protein with canthaxanthin bound | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Chukhutsina, V.U, Baxter, J.M, Fadini, A, Morgan, R.M, Pope, M.A, Maghlaoui, K, Orr, C, Wagner, A, van Thor, J.J. | | Deposit date: | 2022-05-06 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Light activation of Orange Carotenoid Protein reveals bicycle-pedal single-bond isomerization.
Nat Commun, 13, 2022
|
|
7ZSH
 
 | | Structure of Orange Carotenoid Protein with canthaxanthin bound after 2 minutes of illumination | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Chukhutsina, V.U, Baxter, J.M, Fadini, A, Morgan, R.M, Pope, M.A, Maghlaoui, K, Orr, C, Wagner, A, van Thor, J.J. | | Deposit date: | 2022-05-06 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Light activation of Orange Carotenoid Protein reveals bicycle-pedal single-bond isomerization.
Nat Commun, 13, 2022
|
|
7ZSI
 
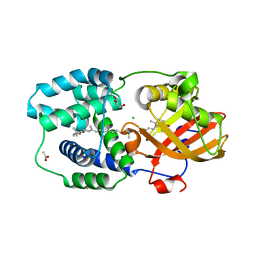 | | Structure of Orange Carotenoid Protein with canthaxanthin bound after 5 minutes of illumination | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Chukhutsina, V.U, Baxter, J.M, Fadini, A, Morgan, R.M, Pope, M.A, Maghlaoui, K, Orr, C, Wagner, A, van Thor, J.J. | | Deposit date: | 2022-05-06 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Light activation of Orange Carotenoid Protein reveals bicycle-pedal single-bond isomerization.
Nat Commun, 13, 2022
|
|
4AIF
 
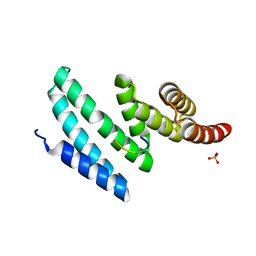 | | AIP TPR domain in complex with human Hsp90 peptide | | Descriptor: | AH RECEPTOR-INTERACTING PROTEIN, HEAT SHOCK PROTEIN HSP 90-ALPHA, SULFATE ION | | Authors: | Morgan, R.M.L, Roe, S.M, Pearl, L.H, Prodromou, C. | | Deposit date: | 2012-02-09 | | Release date: | 2013-01-23 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Structure of the Tpr Domain of Aip: Lack of Client Protein Interaction with the C-Terminal Alpha-7 Helix of the Tpr Domain of Aip is Sufficient for Pituitary Adenoma Predisposition.
Plos One, 7, 2012
|
|
4APO
 
 | | AIP TPR domain in complex with human Tomm20 peptide | | Descriptor: | AH RECEPTOR-INTERACTING PROTEIN, DODECAETHYLENE GLYCOL, MITOCHONDRIAL IMPORT RECEPTOR SUBUNIT TOM20 HOMOLOG, ... | | Authors: | Morgan, R.M.L, Roe, S.M, Pearl, L.H, Prodromou, C. | | Deposit date: | 2012-04-04 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Structure of the Tpr Domain of Aip: Lack of Client Protein Interaction with the C-Terminal Alpha-7 Helix of the Tpr Domain of Aip is Sufficient for Pituitary Adenoma Predisposition.
Plos One, 7, 2012
|
|
4CV4
 
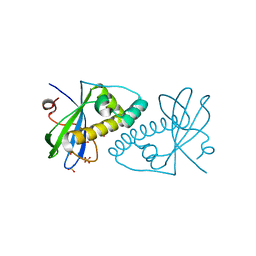 | | PIH N-terminal domain | | Descriptor: | COBALT (II) ION, PIH1 DOMAIN-CONTAINING PROTEIN 1, SULFATE ION | | Authors: | Morgan, R.M, Roe, S.M. | | Deposit date: | 2014-03-23 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structural Basis for Phosphorylation-Dependent Recruitment of Tel2 to Hsp90 by Pih1.
Structure, 22, 2014
|
|
4CSE
 
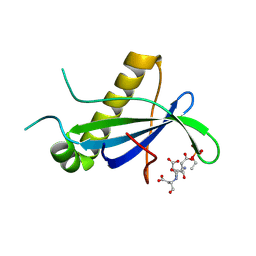 | | PIH N-terminal domain | | Descriptor: | PIH1 DOMAIN-CONTAINING PROTEIN 1, TELOMERE LENGTH REGULATION PROTEIN TEL2 HOMOLOG | | Authors: | Morgan, R.M, Roe, S.M. | | Deposit date: | 2014-03-07 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Basis for Phosphorylation-Dependent Recruitment of Tel2 to Hsp90 by Pih1.
Structure, 22, 2014
|
|
4CKT
 
 | | PIH1 N-terminal domain | | Descriptor: | PIH1 DOMAIN-CONTAINING PROTEIN 1, TELOMERE LENGTH REGULATION PROTEIN TEL2 HOMOLOG | | Authors: | Morgan, R.M, Roe, S.M. | | Deposit date: | 2014-01-08 | | Release date: | 2014-05-14 | | Last modified: | 2014-06-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for Phosphorylation-Dependent Recruitment of Tel2 to Hsp90 by Pih1.
Structure, 22, 2014
|
|
7OPP
 
 | | Crystal structure of the Rab27a fusion with Slp2a-RBDa1 effector for SF4 pocket drug targeting | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Synaptotagmin-like protein 2,Ras-related protein Rab-27A | | Authors: | Jamshidiha, M, Tersa, M, Lanyon-Hogg, T, Perez-Dorado, I, Sutherell, C.L, De Vita, E, Morgan, R.M.L, Tate, E.W, Cota, E. | | Deposit date: | 2021-06-01 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Identification of the first structurally validated covalent ligands of the small GTPase RAB27A.
Rsc Med Chem, 13, 2022
|
|
7OPR
 
 | | Rab27a fusion with Slp2a-RBDa1 effector covalent adduct with CB1 in C123 | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Jamshidiha, M, Tersa, M, Lanyon-Hogg, T, Perez-Dorado, I, Sutherell, C.L, De Vita, E, Morgan, R.M.L, Tate, E.W, Cota, E. | | Deposit date: | 2021-06-01 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Identification of the first structurally validated covalent ligands of the small GTPase RAB27A.
Rsc Med Chem, 13, 2022
|
|
7OPQ
 
 | | Rab27a fusion with Slp2a-RBDa1 effector covalent adduct with CA1 in C188 | | Descriptor: | 1-[(2~{S})-2-(4-methoxyphenyl)pyrrolidin-1-yl]propan-1-one, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Jamshidiha, M, Tersa, M, Lanyon-Hogg, T, Perez-Dorado, I, Sutherell, C.L, De Vita, E, Morgan, R.M.L, Tate, E.W, Cota, E. | | Deposit date: | 2021-06-01 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Identification of the first structurally validated covalent ligands of the small GTPase RAB27A.
Rsc Med Chem, 13, 2022
|
|
8OG4
 
 | | Exostosin-like 3 UDP complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Exostosin-like 3, MANGANESE (II) ION, ... | | Authors: | Sammon, D, Hohenester, E. | | Deposit date: | 2023-03-18 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular mechanism of decision-making in glycosaminoglycan biosynthesis.
Nat Commun, 14, 2023
|
|
8OG1
 
 | | Exostosin-like 3 apo enzyme | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Exostosin-like 3, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Sammon, D, Hohenester, E. | | Deposit date: | 2023-03-17 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Molecular mechanism of decision-making in glycosaminoglycan biosynthesis.
Nat Commun, 14, 2023
|
|
7NJZ
 
 | | X-ray crystallography study of RoAb13 which binds to PIYDIN, a part of the CCR5 N terminal domain | | Descriptor: | Antibody RoAb13 Heavy Chain, Antibody RoAb13 Light Chain, Region from C-C chemokine receptor type 5 N-terminal domain | | Authors: | Helliwell, J.R, Chayen, N, Saridakis, E, Govada, L. | | Deposit date: | 2021-02-17 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray crystallographic studies of RoAb13 bound to PIYDIN, a part of the N-terminal domain of C-C chemokine receptor 5.
Iucrj, 8, 2021
|
|
7NW3
 
 | | X-ray crystallographic study of PIYDIN, which contains the truncation determinants of binding PI and N, bound to RoAb13, a CCR5 antibody | | Descriptor: | Antibody RoAb13 Heavy Chain, Antibody RoAb13 Light Chain, Region from C-C chemokine receptor type 5 N-terminal domain | | Authors: | Saridakis, E, Helliwell, J.R, Govada, L, Chayen, N.E. | | Deposit date: | 2021-03-16 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.200011 Å) | | Cite: | X-ray crystallographic studies of RoAb13 bound to PIYDIN, a part of the N-terminal domain of C-C chemokine receptor 5.
Iucrj, 8, 2021
|
|
6FW7
 
 | | Crystal structure of L-tryptophan oxidase VioA from Chromobacterium violaceum in complex with 4-Fluoro-L-Tryptophan | | Descriptor: | 4-FLUOROTRYPTOPHANE, FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent L-tryptophan oxidase VioA, ... | | Authors: | Lai, H.E, Morgan, M, Moore, S, Freemont, P. | | Deposit date: | 2018-03-05 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A GenoChemetic strategy for derivatization of the violacein natural product scaffold
Biorxiv, 2019
|
|
7QLI
 
 | | Cis structure of rsKiiro at 290 K | | Descriptor: | GLYCEROL, SULFATE ION, rsKiiro | | Authors: | van Thor, J.J, Baxter, J.M. | | Deposit date: | 2021-12-20 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.155 Å) | | Cite: | Optical control of ultrafast structural dynamics in a fluorescent protein.
Nat.Chem., 15, 2023
|
|
7QLL
 
 | |
7QLK
 
 | |
7QLJ
 
 | |
7QLO
 
 | | rsKiiro pump dump probe structure by TR-SFX | | Descriptor: | rsKiiro | | Authors: | van Thor, J.J. | | Deposit date: | 2021-12-20 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Optical control of ultrafast structural dynamics in a fluorescent protein.
Nat.Chem., 15, 2023
|
|
7QLN
 
 | | rsKiiro pump probe structure by TR-SFX | | Descriptor: | rsKiiro | | Authors: | van Thor, J.J. | | Deposit date: | 2021-12-20 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Optical control of ultrafast structural dynamics in a fluorescent protein.
Nat.Chem., 15, 2023
|
|
7QLM
 
 | |
