5L7M
 
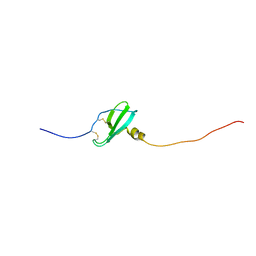 | | Murin CXCL13 solution structure | | Descriptor: | C-X-C motif chemokine 13 | | Authors: | Monneau, Y.R, Lortat-Jacob, H. | | Deposit date: | 2016-06-03 | | Release date: | 2017-06-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of CXCL13 and heparan sulfate binding show that GAG binding site and cellular signalling rely on distinct domains.
Open Biol, 7, 2017
|
|
5IZB
 
 | |
8BXJ
 
 | |
8C2Q
 
 | | Silver ion-bound structure of the silver specific chaperone SilF needed for bacterial silver resistance | | Descriptor: | Copper ABC transporter substrate-binding protein, SILVER ION | | Authors: | Monneau, Y.R, Walker, O, Hologne, M. | | Deposit date: | 2022-12-22 | | Release date: | 2023-10-25 | | Method: | SOLUTION NMR | | Cite: | The battle for silver binding: How the interplay between the SilE, SilF, and SilB proteins contributes to the silver efflux pump mechanism.
J.Biol.Chem., 299, 2023
|
|
6I01
 
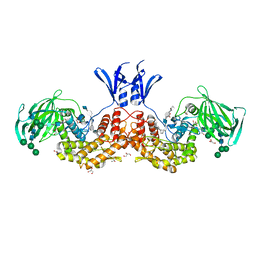 | | Structure of human D-glucuronyl C5 epimerase in complex with substrate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Debarnot, C, Monneau, Y.R, Roig-Zamboni, V, Le Narvor, C, Goulet, A, Fadel, F, Vives, R.R, Bonnaffe, D, Lortat-Jacob, H, Bourne, Y. | | Deposit date: | 2018-10-24 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate binding mode and catalytic mechanism of human heparan sulfate d-glucuronyl C5 epimerase.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6I02
 
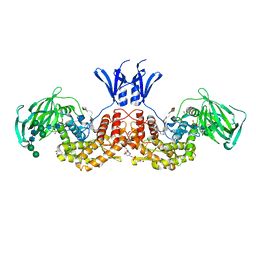 | | Structure of human D-glucuronyl C5 epimerase in complex with product | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Debarnot, C, Monneau, Y.R, Roig-Zamboni, V, Le Narvor, C, Goulet, A, Fadel, F, Vives, R.R, Bonnaffe, D, Lortat-Jacob, H, Bourne, Y. | | Deposit date: | 2018-10-24 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Substrate binding mode and catalytic mechanism of human heparan sulfate d-glucuronyl C5 epimerase.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6HZZ
 
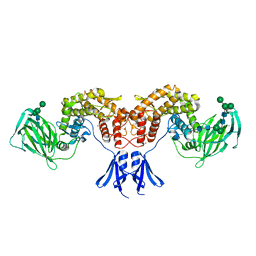 | | Structure of human D-glucuronyl C5 epimerase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Debarnot, C, Monneau, Y.R, Roig-Zamboni, V, Le Narvor, C, Goulet, A, Fadel, F, Vives, R.R, Bonnaffe, D, Lortat-Jacob, H, Bourne, Y. | | Deposit date: | 2018-10-24 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Substrate binding mode and catalytic mechanism of human heparan sulfate d-glucuronyl C5 epimerase.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4BF8
 
 | | Fpr4 PPI domain | | Descriptor: | FPR4 | | Authors: | Monneau, Y, Mackereth, C. | | Deposit date: | 2013-03-15 | | Release date: | 2013-07-31 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure and Activity of the Peptidyl-Prolyl Isomerase Domain from the Histone Chaperone Fpr4 Towards Histone H3 Proline Isomerization
J.Biol.Chem., 288, 2013
|
|
