1YGE
 
 | | LIPOXYGENASE-1 (SOYBEAN) AT 100K | | Descriptor: | FE (III) ION, LIPOXYGENASE-1 | | Authors: | Minor, W, Steczko, J, Stec, B, Otwinowski, Z, Bolin, J.T, Walter, R, Axelrod, B. | | Deposit date: | 1996-06-04 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of soybean lipoxygenase L-1 at 1.4 A resolution.
Biochemistry, 35, 1996
|
|
4XK4
 
 | | E. coli transcriptional regulator RUTR with dihydrouracil | | Descriptor: | DIHYDROPYRIMIDINE-2,4(1H,3H)-DIONE, HTH-type transcriptional regulator RutR | | Authors: | Shumilin, I.A, Cooper, D.R, Shabalin, I.G, Grabowski, M, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-01-09 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | E. COLI TRANSCRIPTIONAL REGULATOR RUTR WITH DIHYDROURACIL
to be published
|
|
6EX7
 
 | | Crystal structure of NDM-1 metallo-beta-lactamase in complex with Cd ions and a hydrolyzed beta-lactam ligand - new refinement | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, CADMIUM ION, ... | | Authors: | Kim, Y, Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A, Tesar, C, Jedrzejczak, R, Babnigg, J, Mire, J, Sacchettini, J, Joachimiak, A. | | Deposit date: | 2017-11-07 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
3KZP
 
 | | Crystal structure of putative diguanylate cyclase/phosphodiesterase from Listaria monocytigenes | | Descriptor: | CACODYLATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Klimecka, M.M, Chruszcz, M, Zimmerman, M.D, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-12-08 | | Release date: | 2009-12-22 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of putative diguanylate cyclase/phosphodiesterase from Listaria monocytigenes
To be Published
|
|
3KZL
 
 | | Crystal structure of BA2930 mutant (H183G) in complex with AcCoA | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETYL COENZYME *A, Aminoglycoside N3-acetyltransferase, ... | | Authors: | Klimecka, M.M, Chruszcz, M, Porebski, P.J, Cymborowski, M, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-12-08 | | Release date: | 2009-12-22 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Analysis of a Putative Aminoglycoside N-Acetyltransferase from Bacillus anthracis.
J.Mol.Biol., 410, 2011
|
|
1QQQ
 
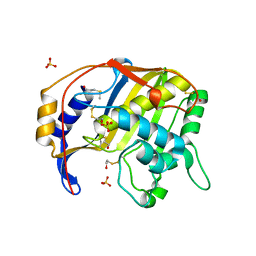 | | CRYSTAL STRUCTURE ANALYSIS OF SER254 MUTANT OF ESCHERICHIA COLI THYMIDYLATE SYNTHASE | | Descriptor: | SULFATE ION, THYMIDYLATE SYNTHASE | | Authors: | Fantz, C, Shaw, D, Jennings, W, Forsthoefel, A, Kitchens, M, Phan, J, Minor, W, Lebioda, L, Berger, F.G, Spencer, H.T. | | Deposit date: | 1999-06-07 | | Release date: | 1999-06-14 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Drug-resistant variants of Escherichia coli thymidylate synthase: effects of substitutions at Pro-254.
Mol.Pharmacol., 57, 2000
|
|
2A35
 
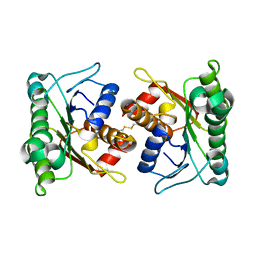 | | 1.5 A Crystal Structure of a Protein of Unknown Function PA4017 from Pseudomonas aeruginosa PAO1, Possible Epimerase | | Descriptor: | hypothetical protein PA4017 | | Authors: | Zhang, R, Xu, L, Cuff, M, Savchenko, A, Cymborowski, M, Minor, W, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-06-23 | | Release date: | 2005-08-09 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5A crystal structure of a hypothetical protein PA4017 from
Pseudomonas aeruginosa PAO1
To be Published
|
|
2AV9
 
 | | Crystal Structure of the PA5185 protein from Pseudomonas Aeruginosa Strain PAO1. | | Descriptor: | SULFATE ION, Thioesterase | | Authors: | Chruszcz, M, Wang, S, Cymborowski, M, Kudritska, M, Evdokimova, E, Edwards, A, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-08-29 | | Release date: | 2005-10-18 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Function-biased choice of additives for optimization of protein crystallization - the case of the putative thioesterase PA5185 from Pseudomonas aeruginosa PAO1.
Cryst.Growth Des., 8, 2008
|
|
3RRO
 
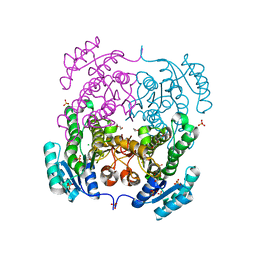 | | Crystal structure of 3-ketoacyl-(acyl-carrier-protein) reductase (FabG) from Vibrio cholerae | | Descriptor: | 1,2-ETHANEDIOL, 3-ketoacyl-(acyl-carrier-protein) reductase, CHLORIDE ION, ... | | Authors: | Hou, J, Chruszcz, M, Cooper, D.R, Grabowski, M, Zheng, H, Osinski, T, Shumilin, I, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-04-29 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dissecting the Structural Elements for the Activation of beta-Ketoacyl-(Acyl Carrier Protein) Reductase from Vibrio cholerae.
J.Bacteriol., 198, 2015
|
|
4XCV
 
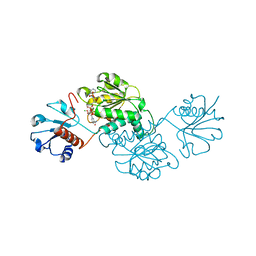 | | Probable 2-hydroxyacid dehydrogenase from Rhizobium etli CFN 42 in complex with NADPH | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, NADP-dependent 2-hydroxyacid dehydrogenase, ... | | Authors: | Langner, K.M, Shabalin, I.G, Handing, K.B, Gasiorowska, O.A, Stead, M, Hillerich, B.S, Chowdhury, S, Hammonds, J, Zimmerman, M.D, Al Obadi, N, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-12-18 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of 2-hydroxyacid dehydrogenase from Rhizobium etli CFN 42 in complex with NADPH
to be published
|
|
4X9K
 
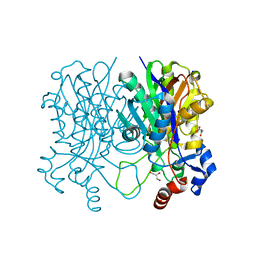 | | Beta-ketoacyl-acyl carrier protein synthase III-2 (FabH2)(C113A) from Vibrio cholerae | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3 protein 2, GLYCEROL, MALONATE ION, ... | | Authors: | Hou, J, Chruszcz, M, Zheng, H, Grabowski, M, Chordia, M.D, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-12-11 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural and enzymatic studies of beta-ketoacyl-(acyl carrier protein) synthase III (FabH) from Vibrio cholerae
to be published
|
|
4XA8
 
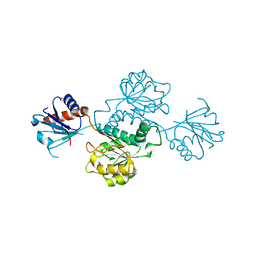 | | Crystal structure of D-isomer specific 2-hydroxyacid dehydrogenase from Xanthobacter autotrophicus Py2 | | Descriptor: | D-isomer specific 2-hydroxyacid dehydrogenase NAD-binding | | Authors: | Handing, K.B, Gasiorowska, O.A, Shabalin, I.G, Cymborowski, M.T, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-12-12 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of D-isomer specific 2-hydroxyacid dehydrogenase from Xanthobacter autotrophicus Py2.
to be published
|
|
4XOX
 
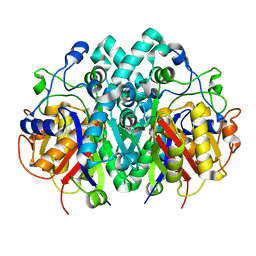 | | Structure of beta-ketoacyl-ACP synthase I (FabB) from Vibrio Cholerae | | Descriptor: | 3-oxoacyl-ACP synthase | | Authors: | Hou, J, Grabowski, M, Cymborowski, M, Zheng, H, Cooper, D.R, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-01-16 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of beta-ketoacyl-ACP synthase I (FabB) from Vibrio Cholerae
to be published
|
|
4YDX
 
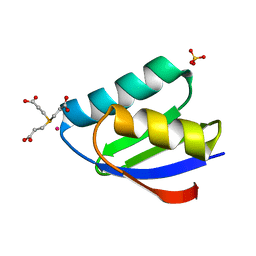 | | Crystal structure of cisplatin bound to a human copper chaperone (monomer) - new refinement | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, Copper transport protein ATOX1, PLATINUM (II) ION, ... | | Authors: | Shabalin, I.G, Boal, A.K, Dauter, Z, Jaskolski, M, Minor, W, Rosenzweig, A.C, Wlodawer, A. | | Deposit date: | 2015-02-23 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Crystallography and chemistry should always go together: a cautionary tale of protein complexes with cisplatin and carboplatin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4YEA
 
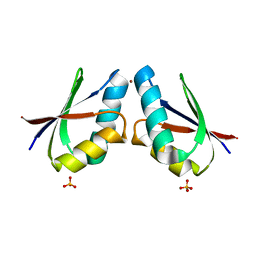 | | Crystal structure of cisplatin bound to a human copper chaperone (dimer) - new refinement | | Descriptor: | COPPER (II) ION, Copper transport protein ATOX1, SULFATE ION | | Authors: | Shabalin, I.G, Dauter, Z, Jaskolski, M, Minor, W, Wlodawer, A. | | Deposit date: | 2015-02-23 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystallography and chemistry should always go together: a cautionary tale of protein complexes with cisplatin and carboplatin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3RRJ
 
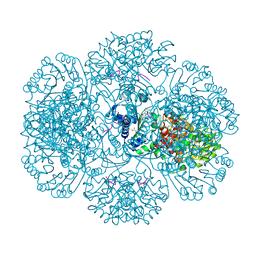 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima in complex with P1,P5-Di(adenosine-5') pentaphosphate | | Descriptor: | BIS(ADENOSINE)-5'-PENTAPHOSPHATE, Bifunctional NAD(P)H-hydrate repair enzyme Nnr, GLYCEROL, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-04-29 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3ROX
 
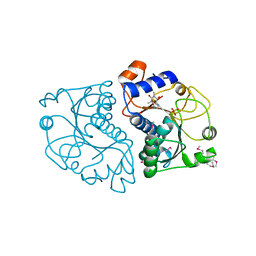 | | Crystal Structure of Mouse Apolipoprotein A-I Binding Protein in Complex with Theophylline | | Descriptor: | Apolipoprotein A-I-binding protein, SULFATE ION, THEOPHYLLINE | | Authors: | Shumilin, I.A, Jha, K.N, Cymborowski, M, Herr, J.C, Minor, W. | | Deposit date: | 2011-04-26 | | Release date: | 2012-07-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RQ5
 
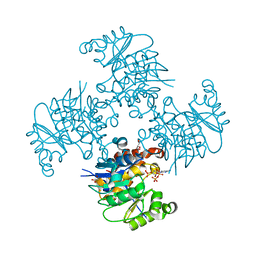 | | Crystal Structure of ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Bacillus subtilis co-crystallized with ATP/Mg2+ and soaked with CoA | | Descriptor: | ADP/ATP-dependent NAD(P)H-hydrate dehydratase, COENZYME A, GLYCEROL | | Authors: | Shumilin, I.A, Cymborowski, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-27 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RQH
 
 | | Crystal Structure of ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Bacillus subtilis in complex with P1,P6-Di(adenosine-5') hexaphosphate | | Descriptor: | ADP/ATP-DEPENDENT NAD(P)H-HYDRATE DEHYDRATASE, MAGNESIUM ION, P1,P6-Di(adenosine-5') hexaphosphate | | Authors: | Shumilin, I.A, Cymborowski, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-28 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RAO
 
 | | Crystal Structure of the Luciferase-like Monooxygenase from Bacillus cereus ATCC 10987. | | Descriptor: | Putative Luciferase-like Monooxygenase, SULFATE ION | | Authors: | Domagalski, M.J, Chruszcz, M, Xu, X, Cui, H, Chin, S, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-03-28 | | Release date: | 2011-05-11 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Luciferase-like Monooxygenase from Bacillus cereus ATCC 10987.
To be Published
|
|
3RT7
 
 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima in complex with ADP-glucose | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE-GLUCOSE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-05-03 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3ROG
 
 | | Crystal Structure of Mouse Apolipoprotein A-I Binding Protein in Complex with Thymidine 3'-monophosphate | | Descriptor: | Apolipoprotein A-I-binding protein, SULFATE ION, THYMIDINE-3'-PHOSPHATE | | Authors: | Shumilin, I.A, Jha, K.N, Cymborowski, M, Herr, J.C, Minor, W. | | Deposit date: | 2011-04-25 | | Release date: | 2012-07-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RQQ
 
 | | Crystal Structure of ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Bacillus subtilis in complex with P1,P3-Di(adenosine-5') triphosphate | | Descriptor: | ADP/ATP-DEPENDENT NAD(P)H-HYDRATE DEHYDRATASE, BIS(ADENOSINE)-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Shumilin, I.A, Cymborowski, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-28 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RS8
 
 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-05-02 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RTE
 
 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima soaked with NADP and ATP. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-05-03 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
