3ZFS
 
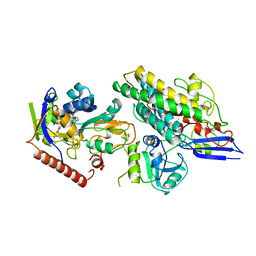 | | Cryo-EM structure of the F420-reducing NiFe-hydrogenase from a methanogenic archaeon with bound substrate | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, COENZYME F420, F420-REDUCING HYDROGENASE, ... | | Authors: | Mills, D.J, Vitt, S, Strauss, M, Shima, S, Vonck, J. | | Deposit date: | 2012-12-12 | | Release date: | 2013-03-06 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | De Novo Modeling of the F420-Reducing [Nife]-Hydrogenase from a Methanogenic Archaeon by Cryo-Electron Microscopy
Elife, 2, 2013
|
|
6F0K
 
 | | Alternative complex III | | Descriptor: | ActD, ActF, ActH, ... | | Authors: | Sousa, J.S, Calisto, F, Mills, D.J, Pereira, M.M, Vonck, J, Kuehlbrandt, W. | | Deposit date: | 2017-11-20 | | Release date: | 2018-05-09 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Structural basis for energy transduction by respiratory alternative complex III.
Nat Commun, 9, 2018
|
|
6F36
 
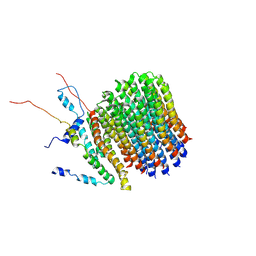 | | Polytomella Fo model | | Descriptor: | Mitochondrial ATP synthase subunit 6, Mitochondrial ATP synthase subunit ASA6, Mitochondrial ATP synthase subunit c | | Authors: | Yildiz, O, Kuehlbrandt, W, Klusch, N, Murphy, B.J, Mills, D.J. | | Deposit date: | 2017-11-28 | | Release date: | 2017-12-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of proton translocation and force generation in mitochondrial ATP synthase.
Elife, 6, 2017
|
|
4V8V
 
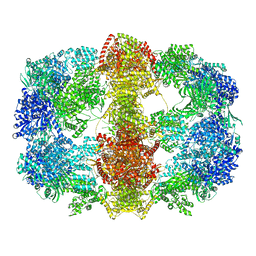 | | Structure and conformational variability of the Mycobacterium tuberculosis fatty acid synthase multienzyme complex | | Descriptor: | FLAVIN MONONUCLEOTIDE, TYPE-I FATTY ACID SYNTHASE | | Authors: | Ciccarelli, L, Connell, S.R, Enderle, M, Mills, D.J, Vonck, J, Grininger, M. | | Deposit date: | 2013-04-18 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Structure and Conformational Variability of the Mycobacterium Tuberculosis Fatty Acid Synthase Multienzyme Complex.
Structure, 21, 2013
|
|
4V8W
 
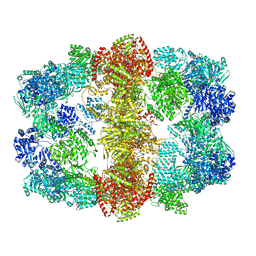 | | Structure and conformational variability of the Mycobacterium tuberculosis fatty acid synthase multienzyme complex | | Descriptor: | FLAVIN MONONUCLEOTIDE, TYPE-I FATTY ACID SYNTHASE | | Authors: | Ciccarelli, L, Connell, S.R, Enderle, M, Mills, D.J, Vonck, J, Grininger, M. | | Deposit date: | 2013-04-18 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (17.5 Å) | | Cite: | Structure and Conformational Variability of the Mycobacterium Tuberculosis Fatty Acid Synthase Multienzyme Complex.
Structure, 21, 2013
|
|
7NXF
 
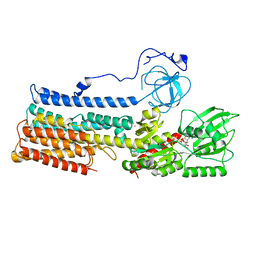 | | Structure of the fungal plasma membrane proton pump Pma1 in its auto-inhibited state - monomer unit | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Heit, S, Geurts, M.M.G, Murphy, B.J, Corey, R, Mills, D.J, Kuehlbrandt, W, Bublitz, M. | | Deposit date: | 2021-03-18 | | Release date: | 2021-11-17 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the hexameric fungal plasma membrane proton pump in its autoinhibited state.
Sci Adv, 7, 2021
|
|
7NY1
 
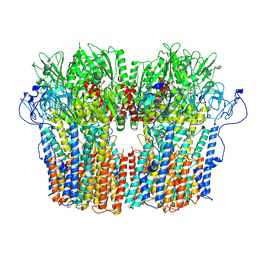 | | Structure of the fungal plasma membrane proton pump Pma1 in its auto-inhibited state - hexameric assembly | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Heit, S, Geurts, M.M.G, Murphy, B.J, Corey, R, Mills, D.J, Kuehlbrandt, W, Bublitz, M. | | Deposit date: | 2021-03-19 | | Release date: | 2021-11-17 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structure of the hexameric fungal plasma membrane proton pump in its autoinhibited state.
Sci Adv, 7, 2021
|
|
5MY1
 
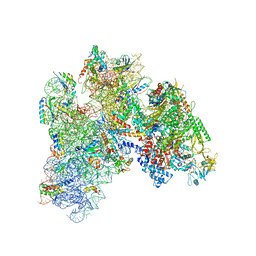 | | E. coli expressome | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Kohler, R, Mooney, R.A, Mills, D.J, Kostrewa, D, Landick, R, Cramer, P. | | Deposit date: | 2017-01-25 | | Release date: | 2017-04-26 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Architecture of a transcribing-translating expressome.
Science, 356, 2017
|
|
5NO4
 
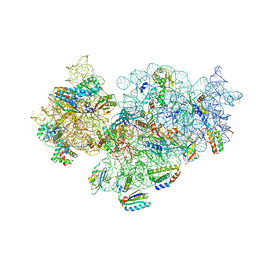 | | RsgA-GDPNP bound to the 30S ribosomal subunit (RsgA assembly intermediate with uS3) | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Lopez-Alonso, J.P, Kaminishi, T, Kikuchi, T, Hirata, Y, Iturrioz, I, Dhimole, N, Schedlbauer, A, Hase, Y, Goto, S, Kurita, D, Muto, A, Zhou, S, Naoe, C, Mills, D.J, Gil-Carton, D, Takemoto, C, Himeno, H, Fucini, P, Connell, S.R. | | Deposit date: | 2017-04-10 | | Release date: | 2017-05-31 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (5.16 Å) | | Cite: | RsgA couples the maturation state of the 30S ribosomal decoding center to activation of its GTPase pocket.
Nucleic Acids Res., 45, 2017
|
|
5NO2
 
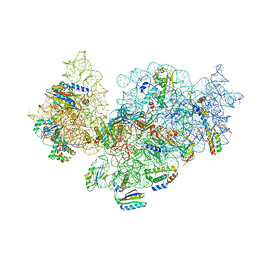 | | RsgA-GDPNP bound to the 30S ribosomal subunit (RsgA assembly intermediate) | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Lopez-Alonso, J.P, Kaminishi, T, Kikuchi, T, Hirata, Y, Iturrioz, I, Dhimole, N, Schedlbauer, A, Hase, Y, Goto, S, Kurita, D, Muto, A, Zhou, S, Naoe, C, Mills, D.J, Gil-Carton, D, Takemoto, C, Himeno, H, Fucini, P, Connell, S.R. | | Deposit date: | 2017-04-10 | | Release date: | 2017-05-24 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (5.16 Å) | | Cite: | RsgA couples the maturation state of the 30S ribosomal decoding center to activation of its GTPase pocket.
Nucleic Acids Res., 45, 2017
|
|
5O8O
 
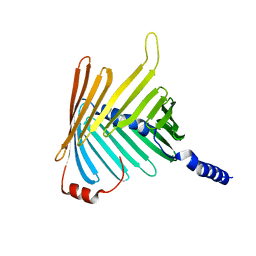 | | N. crassa Tom40 model based on cryo-EM structure of the TOM core complex at 6.8 A | | Descriptor: | Mitochondrial import receptor subunit tom40 | | Authors: | Bausewein, T, Mills, D.J, Nussberger, S, Nitschke, B, Kuehlbrandt, W. | | Deposit date: | 2017-06-13 | | Release date: | 2017-08-16 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Cryo-EM Structure of the TOM Core Complex from Neurospora crassa.
Cell, 170, 2017
|
|
5NO3
 
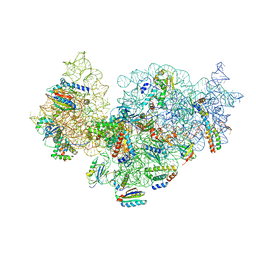 | | RsgA-GDPNP bound to the 30S ribosomal subunit (RsgA assembly intermediate without uS3) | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Lopez-Alonso, J.P, Kaminishi, T, Kikuchi, T, Hirata, Y, Iturrioz, I, Dhimole, N, Schedlbauer, A, Hase, Y, Goto, S, Kurita, D, Muto, A, Zhou, S, Naoe, C, Mills, D.J, Gil-Carton, D, Takemoto, C, Himeno, H, Fucini, P, Connell, S.R. | | Deposit date: | 2017-04-10 | | Release date: | 2017-05-31 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (5.16 Å) | | Cite: | RsgA couples the maturation state of the 30S ribosomal decoding center to activation of its GTPase pocket.
Nucleic Acids Res., 45, 2017
|
|
2YBB
 
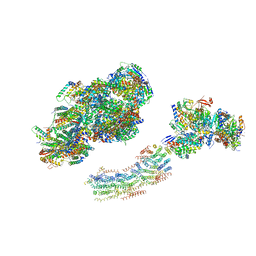 | | Fitted model for bovine mitochondrial supercomplex I1III2IV1 by single particle cryo-EM (EMD-1876) | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, CARDIOLIPIN, ... | | Authors: | Althoff, T, Mills, D.J, Popot, J.-L, Kuehlbrandt, W. | | Deposit date: | 2011-03-02 | | Release date: | 2011-10-19 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (19 Å) | | Cite: | Arrangement of Electron Transport Chain Components in Bovine Mitochondrial Supercomplex I(1)III(2)Iv(1).
Embo J., 30, 2011
|
|
5I68
 
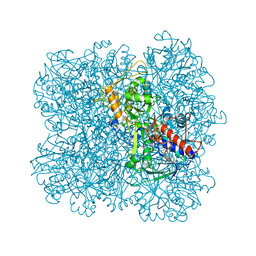 | | Alcohol oxidase from Pichia pastoris | | Descriptor: | Alcohol oxidase 1, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION | | Authors: | Vonck, J, Mills, D.J, Parcej, D.N. | | Deposit date: | 2016-02-16 | | Release date: | 2016-08-03 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structure of Alcohol Oxidase from Pichia pastoris by Cryo-Electron Microscopy.
Plos One, 11, 2016
|
|
5LY6
 
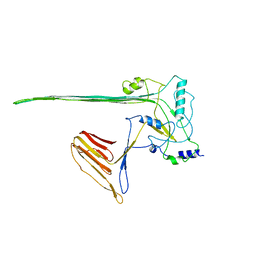 | | CryoEM structure of the membrane pore complex of Pneumolysin at 4.5A | | Descriptor: | Pneumolysin | | Authors: | van Pee, K, Neuhaus, A, D'Imprima, E, Mills, D.J, Kuehlbrandt, W, Yildiz, O. | | Deposit date: | 2016-09-24 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | CryoEM structures of membrane pore and prepore complex reveal cytolytic mechanism of Pneumolysin.
Elife, 6, 2017
|
|
5LUF
 
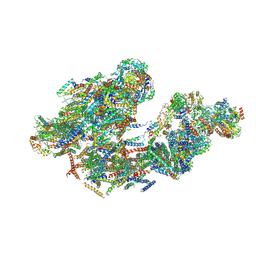 | | Cryo-EM of bovine respirasome | | Descriptor: | COPPER (II) ION, Cytochrome b, Cytochrome b-c1 complex subunit 1, ... | | Authors: | Sousa, J.S, Mills, D.J, Vonck, J, Kuehlbrandt, W. | | Deposit date: | 2016-09-08 | | Release date: | 2016-11-30 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | Functional asymmetry and electron flow in the bovine respirasome.
Elife, 5, 2016
|
|
4CI0
 
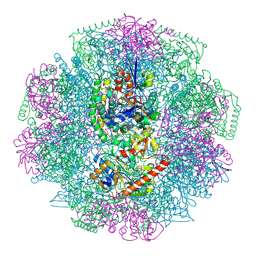 | | Electron cryo-microscopy of F420-reducing NiFe hydrogenase Frh | | Descriptor: | F420-REDUCING HYDROGENASE, SUBUNIT ALPHA, SUBUNIT BETA, ... | | Authors: | Allegretti, M, Mills, D.J, McMullan, G, Kuehlbrandt, W, Vonck, J. | | Deposit date: | 2013-12-05 | | Release date: | 2014-02-26 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Atomic Model of the F420-Reducing [Nife] Hydrogenase by Electron Cryo-Electron Microscopy Using a Direct Electron Detector.
Elife, 3, 2014
|
|
6FKH
 
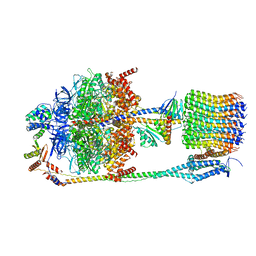 | | Chloroplast F1Fo conformation 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase delta chain, ... | | Authors: | Hahn, A, Vonck, J, Mills, D.J, Meier, T, Kuehlbrandt, W. | | Deposit date: | 2018-01-24 | | Release date: | 2018-05-23 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure, mechanism, and regulation of the chloroplast ATP synthase.
Science, 360, 2018
|
|
6FKI
 
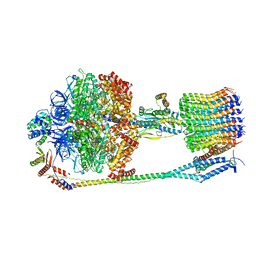 | | Chloroplast F1Fo conformation 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase delta chain, ... | | Authors: | Hahn, A, Vonck, J, Mills, D.J, Meier, T, Kuehlbrandt, W. | | Deposit date: | 2018-01-24 | | Release date: | 2018-05-23 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure, mechanism, and regulation of the chloroplast ATP synthase.
Science, 360, 2018
|
|
6FKF
 
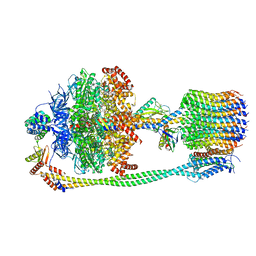 | | Chloroplast F1Fo conformation 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase delta chain, ... | | Authors: | Hahn, A, Vonck, J, Mills, D.J, Meier, T, Kuehlbrandt, W. | | Deposit date: | 2018-01-24 | | Release date: | 2018-05-23 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure, mechanism, and regulation of the chloroplast ATP synthase.
Science, 360, 2018
|
|
6RKD
 
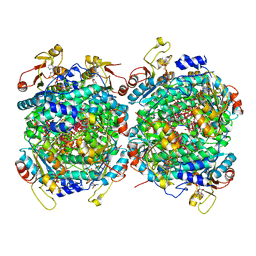 | | Molybdenum storage protein under turnover conditions | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MO(VI)(=O)(OH)2 CLUSTER, ... | | Authors: | Bruenle, S, Mills, D.J, Vonck, J, Ermler, U. | | Deposit date: | 2019-04-30 | | Release date: | 2019-12-18 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molybdate pumping into the molybdenum storage protein via an ATP-powered piercing mechanism.
Proc.Natl.Acad.Sci.USA, 2019
|
|
6S61
 
 | |
4YDS
 
 | |
5AOE
 
 | |
5AOF
 
 | |
