3ZYK
 
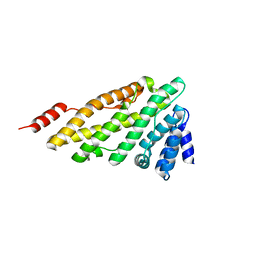 | | Structure of CALM (PICALM) ANTH domain | | Descriptor: | PHOSPHATIDYLINOSITOL-BINDING CLATHRIN ASSEMBLY PROTEIN | | Authors: | Miller, S.E, Sahlender, D.A, Graham, S.C, Honing, S, Robinson, M.S, Peden, A.A, Owen, D.J. | | Deposit date: | 2011-08-23 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The molecular basis for the endocytosis of small R-SNAREs by the clathrin adaptor CALM.
Cell, 147, 2011
|
|
3ZYM
 
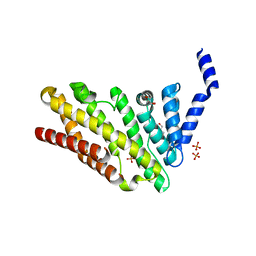 | | Structure of CALM (PICALM) in complex with VAMP8 | | Descriptor: | GLYCEROL, PHOSPHATE ION, PHOSPHATIDYLINOSITOL-BINDING CLATHRIN ASSEMBLY PROTEIN, ... | | Authors: | Miller, S.E, Sahlender, D.A, Graham, S.C, Honing, S, Robinson, M.S, Peden, A.A, Owen, D.J. | | Deposit date: | 2011-08-23 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The molecular basis for the endocytosis of small R-SNAREs by the clathrin adaptor CALM.
Cell, 147, 2011
|
|
3ZYL
 
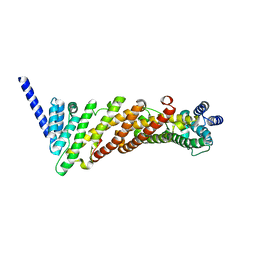 | | Structure of a truncated CALM (PICALM) ANTH domain | | Descriptor: | PHOSPHATIDYLINOSITOL-BINDING CLATHRIN ASSEMBLY PROTEIN | | Authors: | Miller, S.E, Sahlender, D.A, Graham, S.C, Honing, S, Robinson, M.S, Peden, A.A, Owen, D.J. | | Deposit date: | 2011-08-23 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The molecular basis for the endocytosis of small R-SNAREs by the clathrin adaptor CALM.
Cell, 147, 2011
|
|
2QYW
 
 | | Crystal structure of mouse vti1b Habc domain | | Descriptor: | Vesicle transport through interaction with t-SNAREs 1B homolog | | Authors: | Miller, S.E, Collins, B.M, McCoy, A.J, Robinson, M.S, Owen, D.J. | | Deposit date: | 2007-08-15 | | Release date: | 2007-11-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A SNARE-adaptor interaction is a new mode of cargo recognition in clathrin-coated vesicles.
Nature, 450, 2007
|
|
2QY7
 
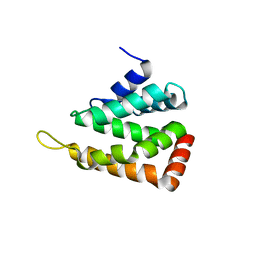 | | Crystal structure of human epsinR ENTH domain | | Descriptor: | Clathrin interactor 1 | | Authors: | Miller, S.E, Collins, B.M, McCoy, A.J, Robinson, M.S, Owen, D.J. | | Deposit date: | 2007-08-13 | | Release date: | 2007-11-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A SNARE-adaptor interaction is a new mode of cargo recognition in clathrin-coated vesicles.
Nature, 450, 2007
|
|
2V8S
 
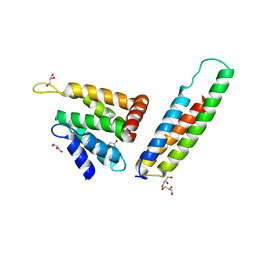 | | VTI1B HABC DOMAIN - EPSINR ENTH DOMAIN COMPLEX | | Descriptor: | CLATHRIN INTERACTOR 1, GLYCEROL, VESICLE TRANSPORT THROUGH INTERACTION WITH T-SNARES HOMOLOG 1B | | Authors: | Owen, D.J, McCoy, A.J, Collins, B.M, Miller, S.E. | | Deposit date: | 2007-08-14 | | Release date: | 2007-11-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | A Snare-Adaptor Interaction is a New Mode of Cargo Recognition in Clathrin Coated Vesicles
Nature, 450, 2007
|
|
1CS7
 
 | | SYNTHETIC DNA HAIRPIN WITH STILBENEDIETHER LINKER | | Descriptor: | 5'-D(GP*(BRU)P*TP*TP*TP*GP*(S02)*CP*AP*AP*AP*AP*C)-3', STRONTIUM ION | | Authors: | Lewis, F.D, Liu, X, Wu, Y, Miller, S.E, Wasielewski, M.R, Letsinger, R.L, Sanishvili, R, Joachimiak, A, Tereshko, V, Egli, M. | | Deposit date: | 1999-08-17 | | Release date: | 2001-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and Photoinduced Electron Transfer in Exceptionally Stable Synthetic DNA Hairpins with Stilbenediether Linkers
J.Am.Chem.Soc., 121, 1999
|
|
8G4Y
 
 | | Structure of ZNRF3 ECD bound to peptide MK1-3.6.10 | | Descriptor: | E3 ubiquitin-protein ligase ZNRF3, MK1-3.6.10 | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2023-02-10 | | Release date: | 2023-12-20 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Potent and selective binders of the E3 ubiquitin ligase ZNRF3 stimulate Wnt signaling and intestinal organoid growth.
Cell Chem Biol, 31, 2024
|
|
8DVN
 
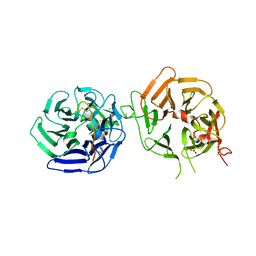 | | Crystal structure of LRP6 E3E4 in complex with disulfide constrained peptide E3.10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Thakur, A.K, Liau, N.P.D, Sudhamsu, J, Hannoush, R.N. | | Deposit date: | 2022-07-29 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Synthetic Multivalent Disulfide-Constrained Peptide Agonists Potentiate Wnt1/ beta-Catenin Signaling via LRP6 Coreceptor Clustering.
Acs Chem.Biol., 18, 2023
|
|
8DVM
 
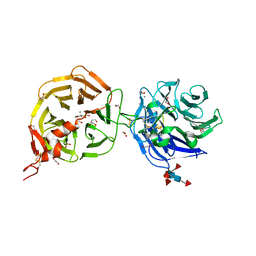 | | Crystal structure of LRP6 E3E4 in complex with disulfide constrained peptide E3.6 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Thakur, A.K, Liau, N.P.D, Sudhamsu, J, Hannoush, R.N. | | Deposit date: | 2022-07-29 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthetic Multivalent Disulfide-Constrained Peptide Agonists Potentiate Wnt1/ beta-Catenin Signaling via LRP6 Coreceptor Clustering.
Acs Chem.Biol., 18, 2023
|
|
8DVL
 
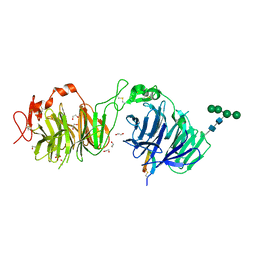 | | Crystal structure of LRP6 E3E4 in complex with disulfide constrained peptide E3.18 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Thakur, A.K, Liau, N.P.D, Sudhamsu, J, Hannoush, R.N. | | Deposit date: | 2022-07-29 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Synthetic Multivalent Disulfide-Constrained Peptide Agonists Potentiate Wnt1/ beta-Catenin Signaling via LRP6 Coreceptor Clustering.
Acs Chem.Biol., 18, 2023
|
|
2JKT
 
 | | AP2 CLATHRIN ADAPTOR CORE with CD4 Dileucine peptide RM(phosphoS) EIKRLLSE Q to E mutant | | Descriptor: | AP-2 COMPLEX SUBUNIT ALPHA-2, AP-2 COMPLEX SUBUNIT BETA-1, AP-2 COMPLEX SUBUNIT MU-1, ... | | Authors: | Owen, D.J, McCoy, A.J, Kelly, B.T, Evans, P.R. | | Deposit date: | 2008-08-29 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | A Structural Explanation for the Binding of Endocytic Dileucine Motifs by the Ap2 Complex.
Nature, 456, 2008
|
|
2JKR
 
 | | AP2 CLATHRIN ADAPTOR CORE with Dileucine peptide RM(phosphoS)QIKRLLSE | | Descriptor: | AP-2 COMPLEX SUBUNIT ALPHA-2, AP-2 COMPLEX SUBUNIT BETA-1, AP-2 COMPLEX SUBUNIT MU-1, ... | | Authors: | Owen, D.J, McCoy, A.J, Kelly, B.T, Evans, P.R. | | Deposit date: | 2008-08-29 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | A Structural Explanation for the Binding of Endocytic Dileucine Motifs by the Ap2 Complex.
Nature, 456, 2008
|
|
