2RAV
 
 | |
2RB5
 
 | |
2RAR
 
 | |
2RBK
 
 | |
3E81
 
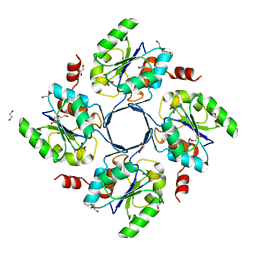 | | Structure-function Analysis of 2-Keto-3-deoxy-D-glycero-D-galacto-nononate-9-phosphate (KDN) Phosphatase Defines a New Clad Within the Type C0 HAD Subfamily | | Descriptor: | 1,2-ETHANEDIOL, Acylneuraminate cytidylyltransferase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lu, Z, Wang, L, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2008-08-19 | | Release date: | 2008-11-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.629 Å) | | Cite: | Structure-Function Analysis of 2-Keto-3-deoxy-D-glycero-D-galactonononate-9-phosphate Phosphatase Defines Specificity Elements in Type C0 Haloalkanoate Dehalogenase Family Members.
J.Biol.Chem., 284, 2009
|
|
3E8M
 
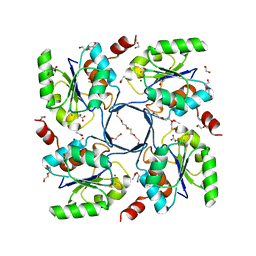 | | Structure-function Analysis of 2-Keto-3-deoxy-D-glycero-D-galacto-nononate-9-phosphate (KDN) Phosphatase Defines a New Clad Within the Type C0 HAD Subfamily | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Acylneuraminate cytidylyltransferase, ... | | Authors: | Lu, Z, Wang, L, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2008-08-20 | | Release date: | 2008-11-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure-Function Analysis of 2-Keto-3-deoxy-D-glycero-D-galactonononate-9-phosphate Phosphatase Defines Specificity Elements in Type C0 Haloalkanoate Dehalogenase Family Members.
J.Biol.Chem., 284, 2009
|
|
3E84
 
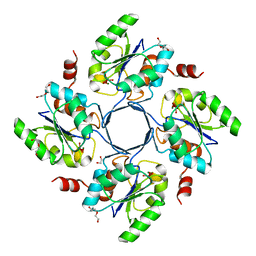 | | Structure-function Analysis of 2-Keto-3-deoxy-D-glycero-D-galacto-nononate-9-phosphate (KDN) Phosphatase Defines a New Clad Within the Type C0 HAD Subfamily | | Descriptor: | 1,2-ETHANEDIOL, Acylneuraminate cytidylyltransferase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lu, Z, Wang, L, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2008-08-19 | | Release date: | 2008-11-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Function Analysis of 2-Keto-3-deoxy-D-glycero-D-galactonononate-9-phosphate Phosphatase Defines Specificity Elements in Type C0 Haloalkanoate Dehalogenase Family Members.
J.Biol.Chem., 284, 2009
|
|
1YMQ
 
 | |
2MA9
 
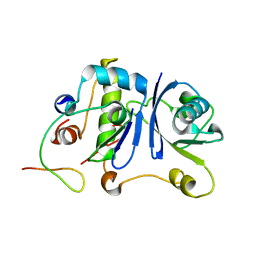 | | HIV-1 Vif SOCS-box and Elongin BC solution structure | | Descriptor: | Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, Virion infectivity factor | | Authors: | Lu, Z, Bergeron, J.R, Atkinson, R.A, Schaller, T, Veselkov, D.A, Oregioni, A, Yang, Y, Matthews, S.J, Malim, M.H, Sanderson, M.R. | | Deposit date: | 2013-07-01 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Insight into the HIV-1 Vif SOCS-box-ElonginBC interaction.
OPEN BIOLOGY, 3, 2013
|
|
4PB9
 
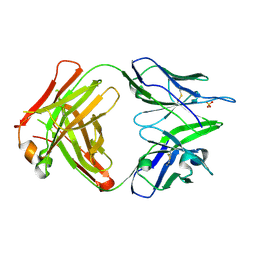 | | Structure of the Fab fragment of the anti-Francisella tularensis GroEL antibody Ab64 | | Descriptor: | Ab64 heavy chain, Ab64 light chain, SULFATE ION | | Authors: | Lu, Z, Rynkiewicz, M.J, Seaton, B.A, Sharon, J. | | Deposit date: | 2014-04-11 | | Release date: | 2014-07-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | B-Cell Epitopes in GroEL of Francisella tularensis.
Plos One, 9, 2014
|
|
4PB0
 
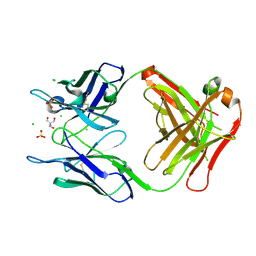 | | Structure of the Fab fragment of the anti-Francisella tularensis GroEL antibody Ab53 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Ab53 heavy chain, Ab53 light chain, ... | | Authors: | Lu, Z, Rynkiewicz, M.J, Seaton, B.A, Sharon, J. | | Deposit date: | 2014-04-10 | | Release date: | 2014-07-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | B-Cell Epitopes in GroEL of Francisella tularensis.
Plos One, 9, 2014
|
|
4OTX
 
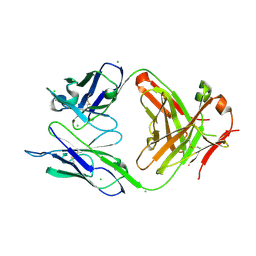 | | Structure of the anti-Francisella tularensis O-antigen antibody N203 Fab fragment | | Descriptor: | AZIDE ION, CHLORIDE ION, N203 heavy chain, ... | | Authors: | Lu, Z, Rynkiewicz, M.J, Yang, C.-Y, Madico, G, Perkins, H.M, Roche, M.I, Seaton, B.A, Sharon, J. | | Deposit date: | 2014-02-14 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional and Structural Characterization of Francisella tularensis O-Antigen Antibodies at the Low End of Antigen Reactivity.
Monoclon Antib Immunodiagn Immunother, 33, 2014
|
|
4KPH
 
 | | Structure of the Fab fragment of N62, a protective monoclonal antibody to the nonreducing end of Francisella tularensis O-antigen | | Descriptor: | ACETATE ION, N62 heavy chain, N62 light chain | | Authors: | Lu, Z, Rynkiewicz, M.J, Yang, C.-Y, Madico, G, Perkins, H.M, Wang, Q, Costello, C.E, Zaia, J, Seaton, B.A, Sharon, J. | | Deposit date: | 2013-05-13 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | The binding sites of monoclonal antibodies to the non-reducing end of Francisella tularensis O-antigen accommodate mainly the terminal saccharide.
Immunology, 140, 2013
|
|
8GTY
 
 | |
3R4C
 
 | | Divergence of Structure and Function Among Phosphatases of the Haloalkanoate (HAD) Enzyme Superfamily: Analysis of BT1666 from Bacteroides thetaiotaomicron | | Descriptor: | Hydrolase, haloacid dehalogenase-like hydrolase, MAGNESIUM ION, ... | | Authors: | Allen, K.N, Lu, Z, Dunaway-Mariano, D. | | Deposit date: | 2011-03-17 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The X-ray crystallographic structure and specificity profile of HAD superfamily phosphohydrolase BT1666: Comparison of paralogous functions in B. thetaiotaomicron.
Proteins, 79, 2011
|
|
8GTZ
 
 | |
2L0K
 
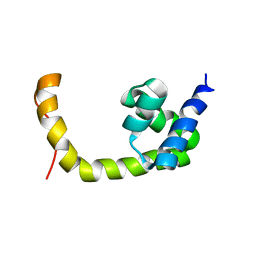 | | NMR solution structure of a transcription factor SpoIIID in complex with DNA | | Descriptor: | Stage III sporulation protein D | | Authors: | Chen, B, Himes, P, Lu, Z, Liu, A, Yan, H, Kroos, L. | | Deposit date: | 2010-07-08 | | Release date: | 2011-08-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Novel Mode of DNA Binding by Bacterial Transcription Factor SpoIIID
To be Published
|
|
3GBS
 
 | | Crystal structure of Aspergillus oryzae cutinase | | Descriptor: | Cutinase 1 | | Authors: | Gosser, Y, Lu, Z, Alemu, G, Li, H, Kong, X, Liu, Z, Montclare, J. | | Deposit date: | 2009-02-20 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and functional studies of Aspergillus oryzae cutinase: enhanced thermostability and hydrolytic activity of synthetic ester and polyester degradation.
J.Am.Chem.Soc., 131, 2009
|
|
4ZLK
 
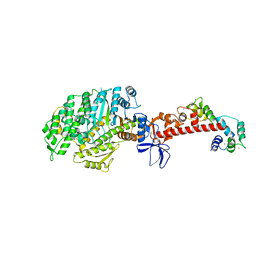 | | Crystal structure of mouse myosin-5a in complex with calcium-bound calmodulin | | Descriptor: | CALCIUM ION, Calmodulin, Unconventional myosin-Va | | Authors: | Shen, M, Zhang, N, Zheng, S, Zhang, W.-B, Zhang, H.-M, Lu, Z, Su, Q.P, Sun, Y, Ye, K, Li, X.-D. | | Deposit date: | 2015-05-01 | | Release date: | 2016-05-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structural basis for calcium regulation of myosin 5 motor function
To Be Published
|
|
1IHZ
 
 | |
1II3
 
 | |
5KZO
 
 | |
1LIR
 
 | | LQ2 FROM LEIURUS QUINQUESTRIATUS, NMR, 22 STRUCTURES | | Descriptor: | LQ2 | | Authors: | Renisio, J.G, Lu, Z, Blanc, E, Jin, W, Lewis, J.H, Bornet, O, Darbon, H. | | Deposit date: | 1998-04-02 | | Release date: | 1998-06-17 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | Solution structure of potassium channel-inhibiting scorpion toxin Lq2.
Proteins, 34, 1999
|
|
1XG3
 
 | | Crystal structure of the C123S 2-methylisocitrate lyase mutant from Escherichia coli in complex with the reaction product, Mg(II)-pyruvate and succinate | | Descriptor: | MAGNESIUM ION, PYRUVIC ACID, Probable methylisocitrate lyase, ... | | Authors: | Liu, S, Lu, Z, Han, Y, Melamud, E, Dunaway-Mariano, D, Herzberg, O. | | Deposit date: | 2004-09-16 | | Release date: | 2005-03-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of 2-Methylisocitrate Lyase in Complex with Product and with Isocitrate Inhibitor Provide Insight into Lyase Substrate Specificity, Catalysis and Evolution
Biochemistry, 44, 2005
|
|
1XG4
 
 | | Crystal Structure of the C123S 2-Methylisocitrate Lyase Mutant from Escherichia coli in complex with the inhibitor isocitrate | | Descriptor: | ISOCITRIC ACID, MAGNESIUM ION, Probable methylisocitrate lyase | | Authors: | Liu, S, Lu, Z, Han, Y, Melamud, E, Dunaway-Mariano, D, Herzberg, O. | | Deposit date: | 2004-09-16 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of 2-Methylisocitrate Lyase in Complex with Product and with Isocitrate Inhibitor Provide Insight into Lyase Substrate Specificity, Catalysis and Evolution
Biochemistry, 44, 2005
|
|
