1OCX
 
 | | E. coli maltose-O-acetyltransferase | | Descriptor: | MALTOSE O-ACETYLTRANSFERASE, TRIMETHYL LEAD ION | | Authors: | Lo Leggio, L, Dal Degan, F, Poulsen, P, Larsen, S. | | Deposit date: | 2003-02-11 | | Release date: | 2003-06-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Structure and Specificity of Escherichia Coli Maltose Acetyltransferase Give New Insight Into the Laca Family of Acyltransferases.
Biochemistry, 42, 2003
|
|
1GOK
 
 | |
1GOQ
 
 | |
1GOM
 
 | |
4OPB
 
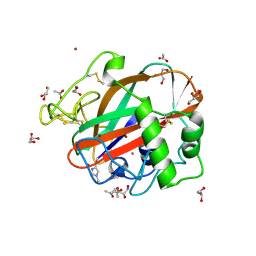 | | AA13 Lytic polysaccharide monooxygenase from Aspergillus oryzae | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, COPPER (II) ION, ... | | Authors: | Lo Leggio, L, Frandsen, K.H, Davies, G.J, Dupree, P, Walton, P, Henrissat, B, Stringer, M, Tovborg, M, De Maria, L, Johansen, K.S. | | Deposit date: | 2014-02-05 | | Release date: | 2015-01-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and boosting activity of a starch-degrading lytic polysaccharide monooxygenase.
Nat Commun, 6, 2015
|
|
1K6A
 
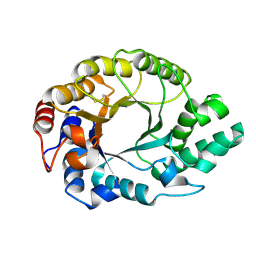 | | Structural studies on the mobility in the active site of the Thermoascus aurantiacus xylanase I | | Descriptor: | xylanase I | | Authors: | Lo Leggio, L, Kalogiannis, S, Eckert, K, Teixeira, S.C.M, Bhat, M.K, Andrei, C, Pickersgill, R.W, Larsen, S. | | Deposit date: | 2001-10-15 | | Release date: | 2002-07-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Substrate specificity and subsite mobility in T. aurantiacus xylanase 10A.
FEBS LETT., 509, 2001
|
|
1GZJ
 
 | |
6H1Z
 
 | | AFGH61B WILD-TYPE | | Descriptor: | ACETATE ION, COPPER (II) ION, Endoglucanase, ... | | Authors: | Lo Leggio, L, Poulsen, J.C.N. | | Deposit date: | 2018-07-12 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structure of a lytic polysaccharide monooxygenase from Aspergillus fumigatus and an engineered thermostable variant.
Carbohydr. Res., 469, 2018
|
|
6HAQ
 
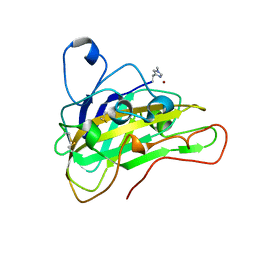 | | AFGH61B WILD-TYPE COPPER LOADED | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, COPPER (II) ION, ... | | Authors: | Lo Leggio, L, Poulsen, J.C.N, Weihe, C.D. | | Deposit date: | 2018-08-08 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structure of a lytic polysaccharide monooxygenase from Aspergillus fumigatus and an engineered thermostable variant.
Carbohydr. Res., 469, 2018
|
|
6HA5
 
 | | AFGH61B L90V/D131S/M134L/A141W VARIANT | | Descriptor: | ACETATE ION, COPPER (II) ION, Endoglucanase, ... | | Authors: | Lo Leggio, L, Poulsen, J.C.N. | | Deposit date: | 2018-08-07 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure of a lytic polysaccharide monooxygenase from Aspergillus fumigatus and an engineered thermostable variant.
Carbohydr. Res., 469, 2018
|
|
1GOR
 
 | |
6F2P
 
 | | Structure of Paenibacillus xanthan lyase to 2.6 A resolution | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Lo Leggio, L, Kadziola, A. | | Deposit date: | 2017-11-27 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Dynamics of a Promiscuous Xanthan Lyase from Paenibacillus nanensis and the Design of Variants with Increased Stability and Activity.
Cell Chem Biol, 26, 2019
|
|
1GOO
 
 | | Thermostable xylanase I from Thermoascus aurantiacus - Cryocooled glycerol complex | | Descriptor: | ENDO-1,4-BETA-XYLANASE, GLYCEROL | | Authors: | Eckert, K, Andrei, C, Larsen, S, Lo Leggio, L. | | Deposit date: | 2001-10-22 | | Release date: | 2001-12-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Substrate Specificity and Subsite Mobility in T. Aurantiacus Xylanase 10A
FEBS Lett., 509, 2001
|
|
4DUL
 
 | | ANAC019 NAC domain crystal form IV | | Descriptor: | NAC domain-containing protein 19 | | Authors: | Lo Leggio, L, Helgstrand, C, Welner, D, Olsen, A.N, Skriver, K. | | Deposit date: | 2012-02-22 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | DNA binding by the plant-specific NAC transcription factors in crystal and solution: a firm link to WRKY and GCM transcription factors.
Biochem.J., 444, 2012
|
|
1E5N
 
 | | E246C mutant of P fluorescens subsp. cellulosa xylanase A in complex with xylopentaose | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE A, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Lo Leggio, L, Jenkins, J.A, Harris, G.W, Pickersgill, R.W. | | Deposit date: | 2000-07-27 | | Release date: | 2000-12-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray crystallographic study of xylopentaose binding to Pseudomonas fluorescens xylanase A.
Proteins, 41, 2000
|
|
6GS0
 
 | | Native Glucuronoyl Esterase from Opitutus terrae | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Putative acetyl xylan esterase, ... | | Authors: | Lo Leggio, L, Larsbrink, J, Meland Knudsen, R, Mazurkewich, S, Navarro Poulsen, J.C. | | Deposit date: | 2018-06-13 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Biochemical and structural features of diverse bacterial glucuronoyl esterases facilitating recalcitrant biomass conversion.
Biotechnol Biofuels, 11, 2018
|
|
6GU8
 
 | | Glucuronoyl Esterase from Solibacter usitatus | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Putative acetyl xylan esterase | | Authors: | Lo Leggio, L, Larsbrink, J, Meland Knudsen, R, Mazurkewich, S, Navarro Poulsen, J.C. | | Deposit date: | 2018-06-19 | | Release date: | 2018-08-15 | | Method: | X-RAY DIFFRACTION (2.01807833 Å) | | Cite: | Biochemical and structural features of diverse bacterial glucuronoyl esterases facilitating recalcitrant biomass conversion.
Biotechnol Biofuels, 11, 2018
|
|
6GRY
 
 | | Glucuronoyl Esterase from Solibacter usitatus. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Putative acetyl xylan esterase, ... | | Authors: | Lo Leggio, L, Larsbrink, J, Meland Knudsen, R, Mazurkewich, S, Navarro Poulsen, J.C. | | Deposit date: | 2018-06-12 | | Release date: | 2018-08-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.0003376 Å) | | Cite: | Biochemical and structural features of diverse bacterial glucuronoyl esterases facilitating recalcitrant biomass conversion.
Biotechnol Biofuels, 11, 2018
|
|
6GRW
 
 | | Glucuronoyl Esterase from Opitutus terrae (Au derivative) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Lo Leggio, L, Larsbrink, J, Meland Knudsen, R, Mazurkewich, S, Navarro Poulsen, J.C. | | Deposit date: | 2018-06-12 | | Release date: | 2018-08-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Biochemical and structural features of diverse bacterial glucuronoyl esterases facilitating recalcitrant biomass conversion.
Biotechnol Biofuels, 11, 2018
|
|
8B4G
 
 | | Structure of a fungal LPMO bound to ligands | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACRYLIC ACID, CHLORIDE ION, ... | | Authors: | Banerjee, S, Huang, Z, Brander, S, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2022-09-20 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.496 Å) | | Cite: | Structure of a fungal LPMO bound to ligands
To Be Published
|
|
1HJS
 
 | | Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BETA-1,4-GALACTANASE, ... | | Authors: | Le Nours, J, Ryttersgaard, C, Lo Leggio, L, Ostergaard, P.R, Borchert, T.V, Christensen, L.L.H, Larsen, S. | | Deposit date: | 2003-02-27 | | Release date: | 2003-06-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure of Two Fungal Beta-1,4-Galactanases: Searching for the Basis for Temperature and Ph Optimum
Protein Sci., 12, 2003
|
|
2XN2
 
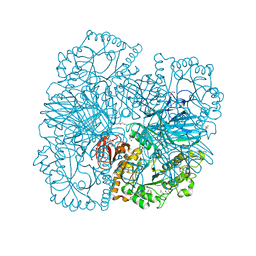 | | Structure of alpha-galactosidase from Lactobacillus acidophilus NCFM with galactose | | Descriptor: | ALPHA-GALACTOSIDASE, GLYCEROL, IMIDAZOLE, ... | | Authors: | Fredslund, F, Abou Hachem, M, Larsen, R.J, Sorensen, P.G, Lo Leggio, L, Svensson, B. | | Deposit date: | 2010-07-30 | | Release date: | 2011-08-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of Alpha-Galactosidase from Lactobacillus Acidophilus Ncfm: Insight Into Tetramer Formation and Substrate Binding.
J.Mol.Biol., 412, 2011
|
|
8QAO
 
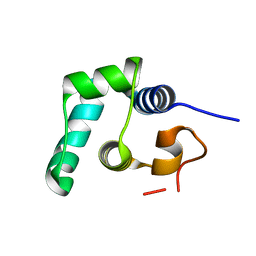 | |
2X2Y
 
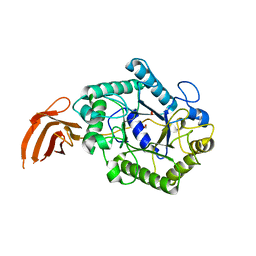 | | Cellulomonas fimi endo-beta-1,4-mannanase double mutant | | Descriptor: | FORMIC ACID, MAGNESIUM ION, MAN26A | | Authors: | Hekmat, O, Lo Leggio, L, Rosengren, A, Kamarauskaite, J, Kolenova, K, Staalbrand, H. | | Deposit date: | 2010-01-18 | | Release date: | 2010-06-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Rational Engineering of Mannosyl Binding in the Distal Glycone Subsites of Cellulomonas Fimi Endo-Beta-1,4-Mannanase: Mannosyl Binding Promoted at Subsite -2 and Demoted at Subsite -3 .
Biochemistry, 49, 2010
|
|
6EKI
 
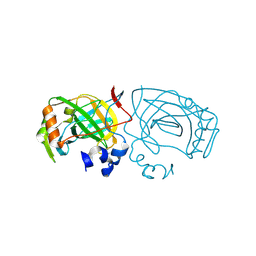 | |
