9YM7
 
 | | Backbone Modification in the Villin Headpiece Miniprotein: HP35 with Calpha-methyl-Phe at Positions 6 and 17, ACPC at Position 29 | | Descriptor: | Villin-1 headpiece: Met12Nle, Phe6alphaMePhe, Phe17alphaMePhe, ... | | Authors: | Lin, Y, David, R.M, Amin, D.M, Osborne, S.W.J, Horne, W.S. | | Deposit date: | 2025-10-09 | | Release date: | 2026-01-07 | | Method: | SOLUTION NMR | | Cite: | Backbone engineering in the hydrophobic core of villin headpiece.
Rsc Chem Biol, 2025
|
|
9YM6
 
 | | Backbone Modification in the Villin Headpiece Miniprotein: HP35 with Calpha-methyl-Phe at Position 17 | | Descriptor: | Villin-1 | | Authors: | Lin, Y, David, R.M, Amin, D.M, Osborne, S.W.J, Horne, W.S. | | Deposit date: | 2025-10-09 | | Release date: | 2026-01-07 | | Method: | SOLUTION NMR | | Cite: | Backbone engineering in the hydrophobic core of villin headpiece.
Rsc Chem Biol, 2025
|
|
9YM0
 
 | | Backbone Modification in the Villin Headpiece Miniprotein: HP35 with ACPC at Position 28 | | Descriptor: | Villin-1 | | Authors: | Lin, Y, David, R.M, Amin, D.M, Osborne, S.W.J, Horne, W.S. | | Deposit date: | 2025-10-09 | | Release date: | 2026-01-07 | | Method: | SOLUTION NMR | | Cite: | Backbone engineering in the hydrophobic core of villin headpiece.
Rsc Chem Biol, 2025
|
|
9YM3
 
 | | Backbone Modification in the Villin Headpiece Miniprotein: HP35 with ACPC at Position 30 | | Descriptor: | Villin-1 | | Authors: | Lin, Y, David, R.M, Amin, D.M, Osborne, S.W.J, Horne, W.S. | | Deposit date: | 2025-10-09 | | Release date: | 2026-01-07 | | Method: | SOLUTION NMR | | Cite: | Backbone engineering in the hydrophobic core of villin headpiece.
Rsc Chem Biol, 2025
|
|
9YM1
 
 | | Backbone Modification in the Villin Headpiece Miniprotein: HP35 with beta3Lys at Position 29 | | Descriptor: | Villin-1 | | Authors: | Lin, Y, David, R.M, Amin, D.M, Osborne, S.W.J, Horne, W.S. | | Deposit date: | 2025-10-09 | | Release date: | 2026-01-07 | | Method: | SOLUTION NMR | | Cite: | Backbone engineering in the hydrophobic core of villin headpiece.
Rsc Chem Biol, 2025
|
|
9YM5
 
 | | Backbone Modification in the Villin Headpiece Miniprotein: HP35 with Calpha-methyl-Phe at Position 6 | | Descriptor: | Villin-1 | | Authors: | Lin, Y, David, R.M, Amin, D.M, Osborne, S.W.J, Horne, W.S. | | Deposit date: | 2025-10-09 | | Release date: | 2026-01-07 | | Method: | SOLUTION NMR | | Cite: | Backbone engineering in the hydrophobic core of villin headpiece.
Rsc Chem Biol, 2025
|
|
9YLZ
 
 | | Backbone Modification in the Villin Headpiece Miniprotein: HP35 with beta3Leu at Position 28 | | Descriptor: | Villin-1 | | Authors: | Lin, Y, David, R.M, Amin, D.M, Osborne, S.W.J, Horne, W.S. | | Deposit date: | 2025-10-09 | | Release date: | 2026-01-07 | | Method: | SOLUTION NMR | | Cite: | Backbone engineering in the hydrophobic core of villin headpiece.
Rsc Chem Biol, 2025
|
|
9YM2
 
 | | Backbone Modification in the Villin Headpiece Miniprotein: HP35 with ACPC at Position 29 | | Descriptor: | Villin-1 | | Authors: | Lin, Y, David, R.M, Amin, D.M, Osborne, S.W.J, Horne, W.S. | | Deposit date: | 2025-10-09 | | Release date: | 2026-01-07 | | Method: | SOLUTION NMR | | Cite: | Backbone engineering in the hydrophobic core of villin headpiece.
Rsc Chem Biol, 2025
|
|
9YM4
 
 | | Backbone Modification in the Villin Headpiece Miniprotein: HP35 with beta3Phe at Position 6 | | Descriptor: | Villin-1 | | Authors: | Lin, Y, David, R.M, Amin, D.M, Osborne, S.W.J, Horne, W.S. | | Deposit date: | 2025-10-09 | | Release date: | 2026-01-07 | | Method: | SOLUTION NMR | | Cite: | Backbone engineering in the hydrophobic core of villin headpiece.
Rsc Chem Biol, 2025
|
|
8FPW
 
 | |
8FPX
 
 | |
6V6M
 
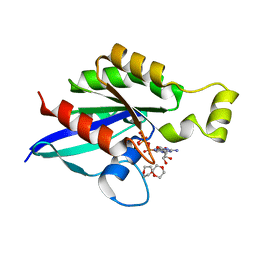 | | Crystal structure of an inactive state of GMPPNP-bound RhoA | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Lin, Y, Zheng, Y. | | Deposit date: | 2019-12-05 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structure of an inactive conformation of GTP-bound RhoA GTPase.
Structure, 29, 2021
|
|
6V6U
 
 | |
6V6V
 
 | |
9JKQ
 
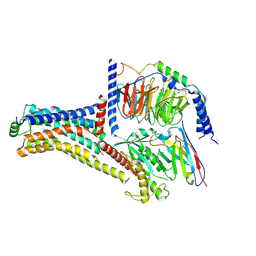 | | Cryo-EM structure of the METH-bound hTAAR1-Gs complex | | Descriptor: | (2S)-N-methyl-1-phenylpropan-2-amine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Lin, Y, Wang, J, Shi, F. | | Deposit date: | 2024-09-16 | | Release date: | 2024-10-16 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Molecular Mechanisms of Methamphetamine-Induced Addiction via TAAR1 Activation.
J.Med.Chem., 67, 2024
|
|
9BB5
 
 | |
9BB7
 
 | |
9BB1
 
 | |
9BB2
 
 | |
9BB6
 
 | |
9BB4
 
 | |
9BB3
 
 | |
5GCN
 
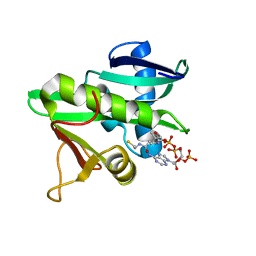 | | CATALYTIC DOMAIN OF TETRAHYMENA GCN5 HISTONE ACETYLTRANSFERASE IN COMPLEX WITH COENZYME A | | Descriptor: | COENZYME A, HISTONE ACETYLTRANSFERASE GCN5 | | Authors: | Lin, Y, Fletcher, C.M, Zhou, J, Allis, C.D, Wagner, G. | | Deposit date: | 1999-03-24 | | Release date: | 1999-07-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the catalytic domain of GCN5 histone acetyltransferase bound to coenzyme A
Nature, 400, 1999
|
|
4GYS
 
 | |
4GYR
 
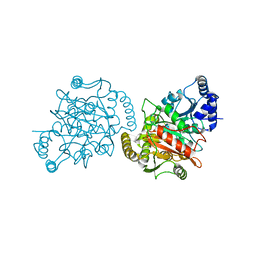 | | Granulibacter bethesdensis allophanate hydrolase apo | | Descriptor: | Allophanate hydrolase | | Authors: | Lin, Y, St Maurice, M. | | Deposit date: | 2012-09-05 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Structure of Allophanate Hydrolase from Granulibacter bethesdensis Provides Insights into Substrate Specificity in the Amidase Signature Family.
Biochemistry, 52, 2013
|
|
