5D39
 
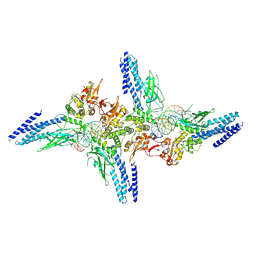 | | Transcription factor-DNA complex | | Descriptor: | DNA (5'-D(P*AP*TP*GP*GP*AP*TP*TP*TP*CP*CP*TP*GP*GP*AP*AP*GP*AP*CP*AP*GP*A)-3'), DNA (5'-D(P*TP*CP*TP*GP*TP*CP*TP*TP*CP*CP*AP*GP*GP*AP*AP*AP*TP*CP*CP*AP*T)-3'), Signal transducer and activator of transcription 6 | | Authors: | Li, J, Niu, F, Ouyang, S, Liu, Z. | | Deposit date: | 2015-08-06 | | Release date: | 2016-08-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for DNA recognition by STAT6
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8I23
 
 | | Clostridium thermocellum RNA polymerase transcription open complex with SigI1 and its promoter | | Descriptor: | DNA (80-mer), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Li, J, Zhang, H, Li, D, Feng, Y, Zhu, P. | | Deposit date: | 2023-01-13 | | Release date: | 2023-10-11 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structure of the transcription open complex of distinct sigma I factors.
Nat Commun, 14, 2023
|
|
8I24
 
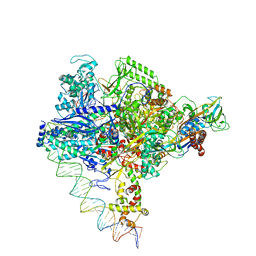 | | Clostridium thermocellum RNA polymerase transcription open complex with SigI6 and its promoter | | Descriptor: | DNA (80-mer), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Li, J, Zhang, H, Li, D, Feng, Y, Zhu, P. | | Deposit date: | 2023-01-13 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structure of the transcription open complex of distinct sigma I factors.
Nat Commun, 14, 2023
|
|
6L8W
 
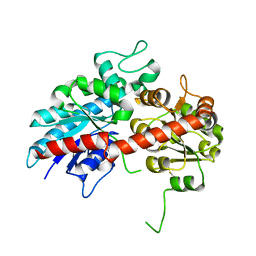 | | Crystal structure of ugt transferase mutant2 | | Descriptor: | Glycosyltransferase | | Authors: | Li, J, Shan, N, Yang, J.G, Liu, W.D, Sun, Y.X. | | Deposit date: | 2019-11-07 | | Release date: | 2020-04-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Efficient O-Glycosylation of Triterpenes Enabled by Protein Engineering of Plant Glycosyltransferase UGT74AC1
Acs Catalysis, 2020
|
|
6L90
 
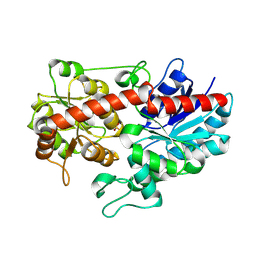 | | Crystal structure of ugt transferase enzyme | | Descriptor: | Glycosyltransferase, SULFATE ION | | Authors: | Li, J, Shan, N, Yang, J.G, Liu, W.D, Sun, Y.X. | | Deposit date: | 2019-11-07 | | Release date: | 2020-04-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Efficient O-Glycosylation of Triterpenes Enabled by Protein Engineering of Plant Glycosyltransferase UGT74AC1
Acs Catalysis, 2020
|
|
6L8X
 
 | | Crystal structure of Siraitia grosvenorii ugt transferase mutant2 | | Descriptor: | Glycosyltransferase | | Authors: | Li, J, Shan, N, Yang, J.G, Liu, W.D, Sun, Y.X. | | Deposit date: | 2019-11-07 | | Release date: | 2020-04-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Efficient O-Glycosylation of Triterpenes Enabled by Protein Engineering of Plant Glycosyltransferase UGT74AC1
Acs Catalysis, 10, 2020
|
|
5XR9
 
 | | Crystal structure of Human Hsp90 with FS6 | | Descriptor: | Heat shock protein HSP 90-alpha, N-[3-[5-bromanyl-2,4-bis(oxidanyl)phenyl]-4-(4-methoxyphenyl)-1,2-oxazol-5-yl]ethanamide | | Authors: | Li, J, Shi, F, Xiong, B, He, J.H. | | Deposit date: | 2017-06-08 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Crystal structure of Human Hsp90 with FS6
To Be Published
|
|
5XRE
 
 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor JX1 | | Descriptor: | Heat shock protein HSP 90-alpha, N-[4-(4-methoxyphenyl)-3-[5-(8-methylquinolin-5-yl)-2,4-bis(oxidanyl)phenyl]-1,2-oxazol-5-yl]ethanamide | | Authors: | Li, J, Shi, F, Xiong, B, He, J.H. | | Deposit date: | 2017-06-08 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.497 Å) | | Cite: | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor JX1
To Be Published
|
|
5XQE
 
 | | Crystal structure of Human Hsp90 with FS3 | | Descriptor: | Heat shock protein HSP 90-alpha, N-[3-[5-chloranyl-2,4-bis(oxidanyl)phenyl]-4-(4-methoxyphenyl)-1,2-oxazol-5-yl]-2-methyl-propanamide | | Authors: | Li, J, Shi, F, Xiong, B, He, J.H. | | Deposit date: | 2017-06-07 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Crystal structure of Human Hsp90 with FS3
To Be Published
|
|
5XRD
 
 | |
5XQD
 
 | | Crystal structure of Human Hsp90 with FS2 | | Descriptor: | Heat shock protein HSP 90-alpha, N-[3-[5-chloranyl-2,4-bis(oxidanyl)phenyl]-4-(4-methoxyphenyl)-1,2-oxazol-5-yl]propanamide | | Authors: | Li, J, Shi, F, Xiong, B, He, J.H. | | Deposit date: | 2017-06-07 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Human Hsp90 with FS2
To Be Published
|
|
5XR5
 
 | | Crystal structure of Human Hsp90 with FS4 | | Descriptor: | Heat shock protein HSP 90-alpha, N-[3-[5-chloranyl-2,4-bis(oxidanyl)phenyl]-4-(4-methoxyphenyl)-1,2-oxazol-5-yl]-2,2-dimethyl-propanamide | | Authors: | Li, J, Shi, F, Xiong, B, He, J.H. | | Deposit date: | 2017-06-07 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Human Hsp90 with FS4
To Be Published
|
|
5XRB
 
 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ5 | | Descriptor: | Heat shock protein HSP 90-alpha, N-[3-[5-chloranyl-2,4-bis(oxidanyl)phenyl]-4-(4-methoxyphenyl)-1,2-oxazol-5-yl]cyclopropanecarboxamide | | Authors: | Li, J, Shi, F, Xiong, B, He, J.H. | | Deposit date: | 2017-06-08 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ5
To Be Published
|
|
4Y7K
 
 | | Structure of an archaeal mechanosensitive channel in closed state | | Descriptor: | Large conductance mechanosensitive channel protein,Riboflavin synthase | | Authors: | Li, J, Liu, Z. | | Deposit date: | 2015-02-15 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Mechanical coupling of the multiple structural elements of the large-conductance mechanosensitive channel during expansion
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4Y7J
 
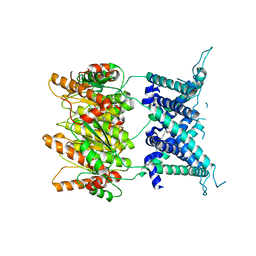 | | Structure of an archaeal mechanosensitive channel in expanded state | | Descriptor: | Large conductance mechanosensitive channel protein,Riboflavin synthase, nonyl beta-D-glucopyranoside | | Authors: | Li, J, Liu, Z. | | Deposit date: | 2015-02-15 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Mechanical coupling of the multiple structural elements of the large-conductance mechanosensitive channel during expansion
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7F04
 
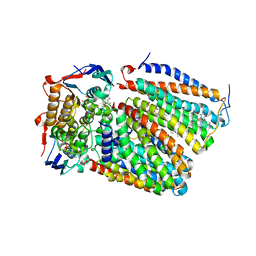 | | Cytochrome c-type biogenesis protein CcmABCD from E. coli in complex with Heme and ATP. | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, ADENOSINE-5'-TRIPHOSPHATE, Cytochrome c biogenesis ATP-binding export protein CcmA, ... | | Authors: | Li, J, Zheng, W, Gu, M, Zhang, K, Zhu, J.P. | | Deposit date: | 2021-06-03 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
4BZ1
 
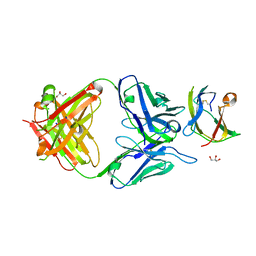 | | Structure of dengue virus EDIII in complex with Fab 3e31 | | Descriptor: | CHLORIDE ION, ENVELOPE PROTEIN, FAB 3E31 HEAVY CHAIN, ... | | Authors: | Li, J, Watterson, D, Chang, C.W, Li, X.Q, Ericsson, D.J, Qiu, L.W, Cai, J.P, Chen, J, Fry, S.R, Cooper, M.A, Che, X.Y, Young, P.R, Kobe, B. | | Deposit date: | 2013-07-23 | | Release date: | 2014-08-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of Dengue Virus Ediii in Complex with Fab 3E31
To be Published
|
|
4BZ2
 
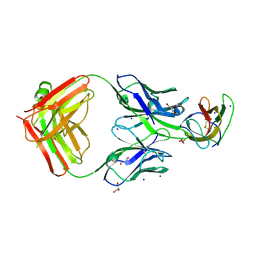 | | Structure of dengue virus EDIII in complex with Fab 2D73 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ENVELOPE PROTEIN, ... | | Authors: | Li, J, Watterson, D, Chang, C.W, Li, X.Q, Ericsson, D.J, Qiu, L.W, Cai, J.P, Chen, J, Fry, S.R, Cooper, M.A, Che, X.Y, Young, P.R, Kobe, B. | | Deposit date: | 2013-07-23 | | Release date: | 2014-08-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure of Dengue Virus Ediii in Complex with Fab 2D73
To be Published
|
|
7FIX
 
 | |
3R93
 
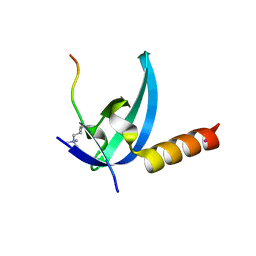 | | Crystal structure of the chromo domain of M-phase phosphoprotein 8 bound to H3K9Me3 peptide | | Descriptor: | H3K9Me3 peptide, M-phase phosphoprotein 8, UNKNOWN ATOM OR ION | | Authors: | Li, J, Li, Z, Ruan, J, Xu, C, Tong, Y, Pan, P.W, Tempel, W, Crombet, L, Min, J, Zang, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-03-24 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.057 Å) | | Cite: | Structural basis for specific binding of human MPP8 chromodomain to histone H3 methylated at lysine 9.
Plos One, 6, 2011
|
|
7F03
 
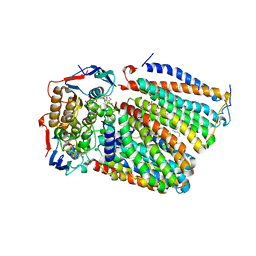 | | Cytochrome c-type biogenesis protein CcmABCD from E. coli in complex with ANP | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, ... | | Authors: | Li, J, Zheng, W, Gu, M, Zhang, K, Zhu, J.P. | | Deposit date: | 2021-06-03 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
7F02
 
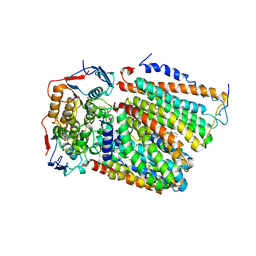 | | Cytochrome c-type biogenesis protein CcmABCD from E. coli | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, ... | | Authors: | Li, J, Zheng, W, Gu, M, Zhang, K, Zhu, J.P. | | Deposit date: | 2021-06-03 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
6LBM
 
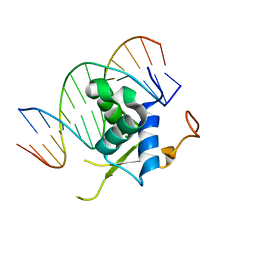 | |
6LBI
 
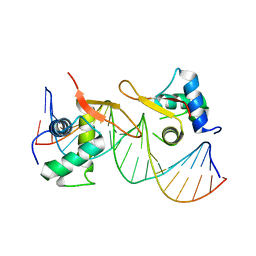 | |
6M3Q
 
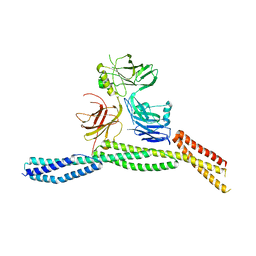 | | Crystal structure of AnkB/beta4-spectrin complex | | Descriptor: | Ankyrin-2, Spectrin beta chain | | Authors: | Li, J, Chen, K, Zhu, R, Zhang, M. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.436 Å) | | Cite: | Structural Basis Underlying Strong Interactions between Ankyrins and Spectrins.
J.Mol.Biol., 432, 2020
|
|
