1A48
 
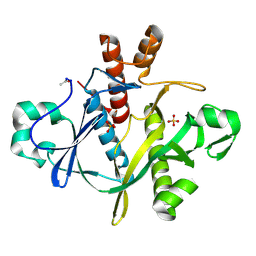 | | SAICAR SYNTHASE | | Descriptor: | PHOSPHORIBOSYLAMINOIMIDAZOLE-SUCCINOCARBOXAMIDE SYNTHASE, SULFATE ION | | Authors: | Levdikov, V.M, Melik-Adamyan, W.R, Lamzin, V.S, Wilson, K.S. | | Deposit date: | 1998-02-12 | | Release date: | 1999-03-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of SAICAR synthase: an enzyme in the de novo pathway of purine nucleotide biosynthesis.
Structure, 6, 1998
|
|
2PFM
 
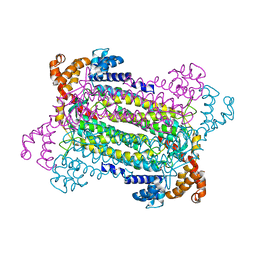 | | Crystal Structure of Adenylosuccinate Lyase (PurB) from Bacillus anthracis | | Descriptor: | Adenylosuccinate lyase, MALONATE ION | | Authors: | Levdikov, V.M, Blagova, E.V, Baumgart, M, Moroz, O.V, Wilkinson, A.J, Wilson, K.S. | | Deposit date: | 2007-04-05 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Adenylosuccinate Lyase (PurB) from Bacillus anthracis
To be Published
|
|
3T9Q
 
 | |
3T91
 
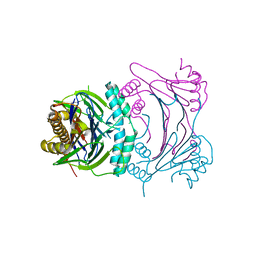 | | Structure of the Phosphatase Domain of the Cell Fate Determinant SpoIIE from Bacillus subtilis | | Descriptor: | MANGANESE (II) ION, Stage II sporulation protein E, alpha-D-mannopyranose, ... | | Authors: | Levdikov, V.M, Blagova, E.V, Wilkinson, A.J. | | Deposit date: | 2011-08-02 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structure of the phosphatase domain of the cell fate determinant SpoIIE from Bacillus subtilis.
J.Mol.Biol., 415, 2012
|
|
3TUF
 
 | |
1T9H
 
 | | The crystal structure of YloQ, a circularly permuted GTPase. | | Descriptor: | ACETATE ION, CALCIUM ION, Probable GTPase engC, ... | | Authors: | Levdikov, V.M, Blagova, E.V, Brannigan, J.A, Cladiere, L, Antson, A.A, Isupov, M.N, Seror, S.J, Wilkinson, A.J. | | Deposit date: | 2004-05-17 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of YloQ, a Circularly Permuted GTPase Essential for Bacillus Subtilis Viability.
J.Mol.Biol., 340, 2004
|
|
1XOC
 
 | | The structure of the oligopeptide-binding protein, AppA, from Bacillus subtilis in complex with a nonapeptide. | | Descriptor: | Nonapeptide VDSKNTSSW, Oligopeptide-binding protein appA, ZINC ION | | Authors: | Levdikov, V.M, Blagova, E.V, Brannigan, J.A, Wright, L, Vagin, A.A, Wilkinson, A.J. | | Deposit date: | 2004-10-06 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The structure of the oligopeptide-binding protein, AppA, from Bacillus subtilis in complex with a nonapeptide.
J.Mol.Biol., 345, 2005
|
|
2B18
 
 | | N-terminal GAF domain of transcriptional pleiotropic repressor CodY. | | Descriptor: | GTP-sensing transcriptional pleiotropic repressor codY, ISOLEUCINE | | Authors: | Levdikov, V.M, Blagova, E, Joseph, P, Sonenshein, A.L, Wilkinson, A.J. | | Deposit date: | 2005-09-15 | | Release date: | 2006-02-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of CodY, a GTP- and Isoleucine-responsive Regulator of Stationary Phase and Virulence in Gram-positive Bacteria.
J.Biol.Chem., 281, 2006
|
|
2B0L
 
 | | C-terminal DNA binding domain of transcriptional pleiotropic repressor CodY. | | Descriptor: | GTP-sensing transcriptional pleiotropic repressor codY | | Authors: | Levdikov, V.M, Blagova, E, Joseph, P, Sonenshein, A.L, Wilkinson, A.J. | | Deposit date: | 2005-09-14 | | Release date: | 2006-02-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Structure of CodY, a GTP- and Isoleucine-responsive Regulator of Stationary Phase and Virulence in Gram-positive Bacteria.
J.Biol.Chem., 281, 2006
|
|
2D30
 
 | | Crystal Structure of Cytidine Deaminase Cdd-2 (BA4525) from Bacillus Anthracis at 2.40A Resolution | | Descriptor: | ZINC ION, cytidine deaminase | | Authors: | Levdikov, V.M, Blagova, E.V, Fogg, M.J, Brannigan, J.A, Moroz, O.V, Wilkinson, A.J, Wilson, K.S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-09-21 | | Release date: | 2006-09-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Cytidine Deaminase Cdd-2 (BA4525) from Bacillus Anthracis at 2.40A Resolution
To be Published
|
|
2GX5
 
 | | N-terminal GAF domain of transcriptional pleiotropic repressor CodY | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, GLYCEROL, GTP-sensing transcriptional pleiotropic repressor codY, ... | | Authors: | Wilkinson, A.J, Levdikov, V.M, Blagova, E.V. | | Deposit date: | 2006-05-08 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The structure of CodY, a GTP- and isoleucine-responsive regulator of stationary phase and virulence in gram-positive bacteria.
J.Biol.Chem., 281, 2006
|
|
2HGV
 
 | |
5LOE
 
 | | Structure of full length Cody from Bacillus subtilis in complex with Ile | | Descriptor: | GTP-sensing transcriptional pleiotropic repressor CodY, ISOLEUCINE | | Authors: | Wilkinson, A.J, Levdikov, V.M, Blagova, E.V. | | Deposit date: | 2016-08-09 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the Branched-chain Amino Acid and GTP-sensing Global Regulator, CodY, from Bacillus subtilis.
J. Biol. Chem., 292, 2017
|
|
5LOJ
 
 | |
5LNH
 
 | | Structure of full length Unliganded CodY from Bacillus subtilis | | Descriptor: | GTP-sensing transcriptional pleiotropic repressor CodY, SULFATE ION | | Authors: | Wilkinson, A.J, Levdikov, V.M, Blagova, E.V. | | Deposit date: | 2016-08-04 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the Branched-chain Amino Acid and GTP-sensing Global Regulator, CodY, from Bacillus subtilis.
J. Biol. Chem., 292, 2017
|
|
5LOO
 
 | |
5MQH
 
 | |
5N0L
 
 | | The structure of the cofactor binding GAF domain of the nutrient sensor CodY from Clostridium difficile | | Descriptor: | GTP-sensing transcriptional pleiotropic repressor CodY, ISOLEUCINE | | Authors: | Levdikov, V.M, Blagova, E.V, Wilkinson, A.J, Sonenshein, A.L. | | Deposit date: | 2017-02-03 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Impact of CodY protein on metabolism, sporulation and virulence in Clostridioides difficile ribotype 027.
Plos One, 14, 2019
|
|
3O6Q
 
 | | The Structure of SpoIISA and SpoIISB, a Toxin - Antitoxin System | | Descriptor: | Stage II sporulation protein SA, Stage II sporulation protein SB | | Authors: | Levdikov, V.M, Blagova, E.V, Lebedev, A.A, Wilkinson, A.J, Florek, P, Barak, I. | | Deposit date: | 2010-07-29 | | Release date: | 2010-12-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure and interactions of SpoIISA and SpoIISB, a toxin-antitoxin system in Bacillus subtilis.
J.Biol.Chem., 161, 2010
|
|
2X36
 
 | | Structure of the proteolytic domain of the Human Mitochondrial Lon protease | | Descriptor: | LON PROTEASE HOMOLOG, MITOCHONDRIAL | | Authors: | Garcia, J, Ondrovicova, G, Blagova, E, Levdikov, V.M, Bauer, J.A, Kutejova, E, Wilkinson, A.J, Wilson, K.S. | | Deposit date: | 2010-01-21 | | Release date: | 2010-05-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Catalytic Domain of the Human Mitochondrial Lon Protease: Proposed Relation of Oligomer Formation and Activity.
Protein Sci., 19, 2010
|
|
2YAL
 
 | | SinR, Master Regulator of biofilm formation in Bacillus subtilis | | Descriptor: | HTH-TYPE TRANSCRIPTIONAL REGULATOR SINR, NICKEL (II) ION | | Authors: | Colledge, V.L, Fogg, M.J, Levdikov, V.M, Leech, A, Dodson, E.J, Wilkinson, A.J. | | Deposit date: | 2011-02-23 | | Release date: | 2011-06-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structure and Organisation of Sinr, the Master Regulator of Biofilm Formation in Bacillus Subtilis.
J.Mol.Biol., 411, 2011
|
|
4ZJN
 
 | | Crystal structure of the bacteriophage G20C portal protein | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Portal protein | | Authors: | Williams, L.S, Turkenburg, J.P, Levdikov, V.M, Minakhin, L, Severinov, K, Antson, A.A. | | Deposit date: | 2015-04-29 | | Release date: | 2015-05-27 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure of the bacteriophage G20C portal protein
To be published
|
|
3DD6
 
 | | Crystal structure of Rph, an exoribonuclease from Bacillus anthracis at 1.7 A resolution | | Descriptor: | Ribonuclease PH, SULFATE ION | | Authors: | Rawlings, A.E, Blagova, E.V, Levdikov, V.M, Fogg, M.J, Wilson, K.S, Wilkinson, A.J, Structural Proteomics in Europe 2 (SPINE-2) | | Deposit date: | 2008-06-05 | | Release date: | 2009-02-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | The structure of Rph, an exoribonuclease from Bacillus anthracis, at 1.7 A resolution.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3O9P
 
 | | The structure of the Escherichia coli murein tripeptide binding protein MppA | | Descriptor: | L-ALA-GAMMA-D-GLU-MESO-DIAMINOPIMELIC ACID, Periplasmic murein peptide-binding protein, ZINC ION | | Authors: | Maqbool, A, Levdikov, V.M, Blagova, E.V, Wilkinson, A.J, Thomas, G.H. | | Deposit date: | 2010-08-04 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Compensating Stereochemical Changes Allow Murein Tripeptide to Be Accommodated in a Conventional Peptide-binding Protein.
J.Biol.Chem., 286, 2011
|
|
3QQ6
 
 | | The N-terminal DNA binding domain of SinR from Bacillus subtilis | | Descriptor: | HTH-type transcriptional regulator sinR | | Authors: | Colledge, V, Fogg, M.J, Levdikov, V.M, Dodson, E.J, Wilkinson, A.J. | | Deposit date: | 2011-02-15 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Organisation of SinR, the Master Regulator of Biofilm Formation in Bacillus subtilis.
J.Mol.Biol., 411, 2011
|
|
