1YBZ
 
 | | Conserved hypothetical protein from Pyrococcus furiosus Pfu-1581948-001 | | Descriptor: | UNKNOWN ATOM OR ION, chorismate mutase | | Authors: | Lee, D, Chen, L, Nguyen, D, Dillard, B.D, Tempel, W, Habel, J, Zhou, W, Chang, S.-H, Kelley, L.-L.C, Liu, Z.-J, Lin, D, Zhang, H, Praissman, J, Bridger, S, Eneh, J.C, Hopkins, R.C, Jenney Jr, F.E, Lee, H.-S, Li, T, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-12-21 | | Release date: | 2005-02-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Conserved hypothetical protein from Pyrococcus furiosus Pfu-1581948-001
To be published
|
|
1LS8
 
 | | NMR structure of the unliganded Bombyx mori pheromone-binding protein at physiological pH | | Descriptor: | pheromone binding protein | | Authors: | Lee, D, Damberger, F, Horst, R, Guntert, P, Leal, W.S, Wuthrich, K. | | Deposit date: | 2002-05-17 | | Release date: | 2002-11-20 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the unliganded Bombyx mori pheromone-binding protein at physiological pH
FEBS Lett., 531, 2002
|
|
2P01
 
 | | The structure of receptor-associated protein(RAP) | | Descriptor: | Alpha-2-macroglobulin receptor-associated protein | | Authors: | Lee, D, Walsh, J.D, Migliorini, M, Yu, P, Cai, T, Schwieters, C.D, Krueger, S, Strickland, D.K, Wang, Y.X. | | Deposit date: | 2007-02-28 | | Release date: | 2007-08-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of receptor-associated protein (RAP).
Protein Sci., 16, 2007
|
|
2P03
 
 | | The structure of receptor-associated protein(RAP) | | Descriptor: | Alpha-2-macroglobulin receptor-associated protein | | Authors: | Lee, D, Walsh, J.D, Migliorini, M, Yu, P, Cai, T, Schwieters, C.D, Krueger, S, Strickland, D.K, Wang, Y.X. | | Deposit date: | 2007-02-28 | | Release date: | 2007-08-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of receptor-associated protein (RAP).
Protein Sci., 16, 2007
|
|
2H8W
 
 | | Solution structure of ribosomal protein L11 | | Descriptor: | 50S ribosomal protein L11 | | Authors: | Lee, D, Walsh, J.D, Yu, P, Choli-Papadopoulou, T, Krueger, S, Draper, D, Wang, Y.-X. | | Deposit date: | 2006-06-08 | | Release date: | 2007-02-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Structure of Free L11 and Functional Dynamics of L11 in Free, L11-rRNA(58 nt) Binary and L11-rRNA(58 nt)-thiostrepton Ternary Complexes.
J.Mol.Biol., 367, 2007
|
|
2FTU
 
 | | solution structure of domain 3 of RAP | | Descriptor: | Alpha-2-macroglobulin receptor-associated protein, domain 3 | | Authors: | Lee, D, Walsh, J.D, Wang, Y.-X. | | Deposit date: | 2006-01-24 | | Release date: | 2006-05-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | RAP uses a histidine switch to regulate its interaction with LRP in the ER and Golgi.
Mol.Cell, 22, 2006
|
|
5ZAI
 
 | |
2E34
 
 | | L11 structure with RDC and RG refinement | | Descriptor: | 50S ribosomal protein L11 | | Authors: | Lee, D, Walsh, J.D, Yu, P, Markus, M.A, Choli-Papadopoulous, T, Schwieters, C.D, Krueger, S, Draper, D.E, Wang, Y.X. | | Deposit date: | 2006-11-20 | | Release date: | 2007-06-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The structure of free L11 and functional dynamics of L11 in free, L11-rRNA(58 nt) binary and L11-rRNA(58 nt)-thiostrepton ternary complexes
J.Mol.Biol., 367, 2007
|
|
2E36
 
 | | L11 with SANS refinement | | Descriptor: | 50S ribosomal protein L11 | | Authors: | Lee, D, Walsh, J.D, Yu, P, Markus, M.A, Choli-Papadopoulous, T, Schwieters, C.D, Krueger, S, Draper, D.E, Wang, Y.X. | | Deposit date: | 2006-11-20 | | Release date: | 2007-06-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The structure of free L11 and functional dynamics of L11 in free, L11-rRNA(58 nt) binary and L11-rRNA(58 nt)-thiostrepton ternary complexes
J.Mol.Biol., 367, 2007
|
|
2E35
 
 | | the minimized average structure of L11 with rg refinement | | Descriptor: | 50S ribosomal protein L11 | | Authors: | Lee, D, Walsh, J.D, Yu, P, Markus, M.A, Choli-Papadopoulous, T, Schwieters, C.D, Krueger, S, Draper, D.E, Wang, Y.X. | | Deposit date: | 2006-11-20 | | Release date: | 2007-06-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The structure of free L11 and functional dynamics of L11 in free, L11-rRNA(58 nt) binary and L11-rRNA(58 nt)-thiostrepton ternary complexes
J.Mol.Biol., 367, 2007
|
|
6IHD
 
 | | Crystal structure of Malate dehydrogenase from Metallosphaera sedula | | Descriptor: | 2-ETHOXYETHANOL, Malate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Lee, D, Kim, K.J. | | Deposit date: | 2018-09-29 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and biochemical characterization of malate dehydrogenase from Metallosphaera sedula
Biochem. Biophys. Res. Commun., 509, 2019
|
|
6IHE
 
 | |
6JX2
 
 | | Crystal structure of Ketol-acid reductoisomerase from Corynebacterium glutamicum | | Descriptor: | 1,2-ETHANEDIOL, Ketol-acid reductoisomerase (NADP(+)), MAGNESIUM ION, ... | | Authors: | Lee, D, Hong, J, Kim, K.-J. | | Deposit date: | 2019-04-22 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure and Biochemical Characterization of Ketol-Acid Reductoisomerase fromCorynebacterium glutamicum.
J.Agric.Food Chem., 67, 2019
|
|
1EJP
 
 | | SOLUTION STRUCTURE OF THE SYNDECAN-4 WHOLE CYTOPLASMIC DOMAIN | | Descriptor: | SYNDECAN-4 | | Authors: | Lee, D, Oh, E.S, Woods, A, Couchman, J.R, Lee, W. | | Deposit date: | 2000-03-03 | | Release date: | 2001-09-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the dimeric cytoplasmic domain of syndecan-4.
Biochemistry, 40, 2001
|
|
2MWH
 
 | | NMR solution structure of ligand-free OAA | | Descriptor: | Anti-HIV lectin OAA | | Authors: | Lee, D, Carneiro, M.G, Koharudin, L.M, Griesinger, C, Gronenborn, A.M, Ban, D, Sabo, T, Trigo-Mourino, P, Mazur, A. | | Deposit date: | 2014-11-10 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Sampling of Glycan-Bound Conformers by the Anti-HIV Lectin Oscillatoria agardhii agglutinin in the Absence of Sugar.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6KHQ
 
 | |
6KGZ
 
 | | bacterial cystathionine gamma-lyase MccB of Staphylococcus aureus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cystathionine gamma-lyase | | Authors: | Ha, N.-C, Lee, D. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Bacterial Cystathionine Gamma-Lyase in The Cysteine Biosynthesis Pathway of Staphylococcus aureus
Crystals, 9, 2019
|
|
3K94
 
 | | Crystal Structure of Thiamin pyrophosphokinase from Geobacillus thermodenitrificans, Northeast Structural Genomics Consortium Target GtR2 | | Descriptor: | Thiamin pyrophosphokinase | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Janjua, J, Xiao, R, Patel, D.J, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-15 | | Release date: | 2010-02-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Northeast Structural Genomics Consortium Target GtR2
To be Published
|
|
3JVC
 
 | | Crystal Structure of the Lipoprotein_17 domain from Q9PRA0_UREPA protein of Ureaplasma parvum. Northeast Structural Genomics Consortium Target UuR17a. | | Descriptor: | Conserved hypothetical membrane lipoprotein | | Authors: | Vorobiev, S, Neely, H, Lee, D, Ciccosanti, C, Mao, L, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-09-16 | | Release date: | 2009-09-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystal Structure of the Lipoprotein_17 domain from Q9PRA0_UREPA protein of Ureaplasma parvum.
To be Published
|
|
4UN2
 
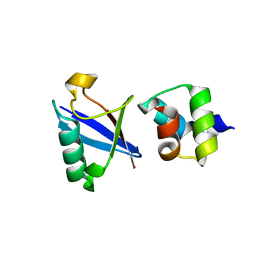 | | Crystal structure of the UBA domain of Dsk2 in complex with Ubiquitin | | Descriptor: | UBIQUITIN, UBIQUITIN DOMAIN-CONTAINING PROTEIN DSK2 | | Authors: | Michielssens, S, Peters, J.H, Ban, D, Pratihar, S, Seeliger, D, Sharma, M, Giller, K, Sabo, T.M, Becker, S, Lee, D, Griesinger, C, de Groot, B.L. | | Deposit date: | 2014-05-23 | | Release date: | 2014-08-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | A Designed Conformational Shift to Control Protein Binding Specificity.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3D5N
 
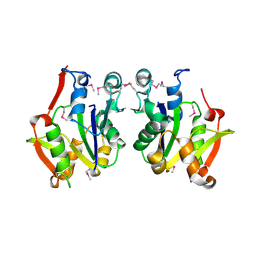 | | Crystal structure of the Q97W15_SULSO protein from Sulfolobus solfataricus. NESG target SsR125. | | Descriptor: | Q97W15_SULSO | | Authors: | Vorobiev, S.M, Chen, Y, Seetharaman, J, Lee, D, Foote, R.E, Maglaqui, M, Janjua, H, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-05-16 | | Release date: | 2008-07-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the Q97W15_SULSO protein from Sulfolobus solfataricus. NESG target SsR125.
To be Published
|
|
3D3Q
 
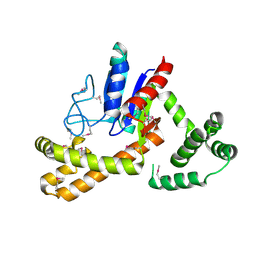 | | Crystal structure of tRNA delta(2)-isopentenylpyrophosphate transferase (SE0981) from Staphylococcus epidermidis. Northeast Structural Genomics Consortium target SeR100 | | Descriptor: | tRNA delta(2)-isopentenylpyrophosphate transferase | | Authors: | Forouhar, F, Abashidze, M, Seetharaman, J, Mao, L, Xiao, R, Maglaqui, M, Lee, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-05-12 | | Release date: | 2008-07-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of tRNA delta(2)-isopentenylpyrophosphate transferase (SE0981) from Staphylococcus epidermidis.
To be Published
|
|
7ER6
 
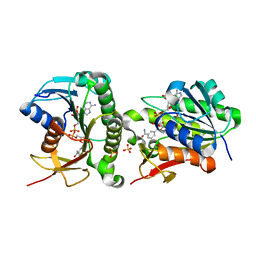 | | Crystal structure of human Biliverdin IX-beta reductase B | | Descriptor: | Flavin reductase (NADPH), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Griesinger, C, Lee, D, Ryu, K.S, Kim, M, Ha, J.H. | | Deposit date: | 2021-05-06 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Repositioning Food and Drug Administration-Approved Drugs for Inhibiting Biliverdin IX beta Reductase B as a Novel Thrombocytopenia Therapeutic Target.
J.Med.Chem., 65, 2022
|
|
7ER9
 
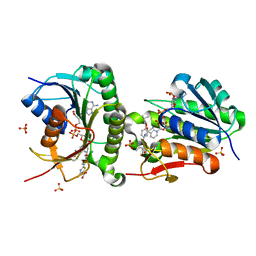 | | Crystal structure of human Biliverdin IX-beta reductase B with Febuxostat (TEI) | | Descriptor: | 2-(3-CYANO-4-ISOBUTOXY-PHENYL)-4-METHYL-5-THIAZOLE-CARBOXYLIC ACID, Flavin reductase (NADPH), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Griesinger, C, Lee, D, Ryu, K.S, Kim, M, Ha, J.H. | | Deposit date: | 2021-05-06 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Repositioning Food and Drug Administration-Approved Drugs for Inhibiting Biliverdin IX beta Reductase B as a Novel Thrombocytopenia Therapeutic Target.
J.Med.Chem., 65, 2022
|
|
7ERE
 
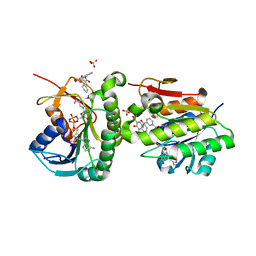 | | Crystal structure of human Biliverdin IX-beta reductase B with Pyrantel Pamoate (PPA) | | Descriptor: | 4-[(3-carboxy-2-oxidanyl-naphthalen-1-yl)methyl]-3-oxidanyl-naphthalene-2-carboxylic acid, Flavin reductase (NADPH), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Griesinger, C, Lee, D, Ryu, K.S, Kim, M, Ha, J.H. | | Deposit date: | 2021-05-06 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Repositioning Food and Drug Administration-Approved Drugs for Inhibiting Biliverdin IX beta Reductase B as a Novel Thrombocytopenia Therapeutic Target.
J.Med.Chem., 65, 2022
|
|
