1O8Q
 
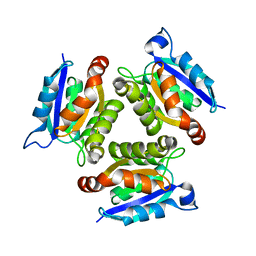 | | The active site of the molybdenum cofactor biosenthetic protein domain Cnx1G | | Descriptor: | MOLYBDOPTERIN BIOSYNTHESIS CNX1 PROTEIN | | Authors: | Kuper, J, Winking, J, Hecht, H.J, Schwarz, G, Mendel, R.R. | | Deposit date: | 2002-11-28 | | Release date: | 2003-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Active Site of the Molybdenum Cofactor Biosynthetic Protein Domain Cnx1G
Arch.Biochem.Biophys., 411, 2003
|
|
1O8N
 
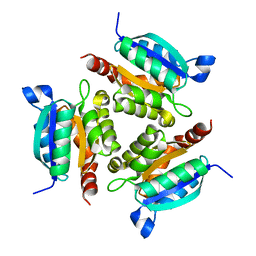 | | The active site of the molybdenum cofactor biosynthetic protein domain Cnx1G | | Descriptor: | MOLYBDOPTERIN BIOSYNTHESIS CNX1 PROTEIN | | Authors: | Kuper, J, Winking, J, Hecht, H.J, Schwarz, G, Mendel, R.R. | | Deposit date: | 2002-11-28 | | Release date: | 2003-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Active Site of the Molybdenum Cofactor Biosynthetic Protein Domain Cnx1G
Arch.Biochem.Biophys., 411, 2003
|
|
1O8O
 
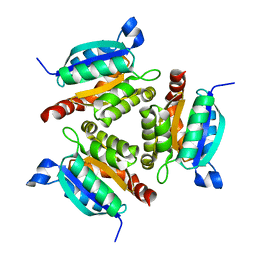 | | The active site of the molybdenum cofactor biosynthetic protein domain Cnx1G | | Descriptor: | MOLYBDOPTERIN BIOSYNTHESIS CNX1 PROTEIN | | Authors: | Kuper, J, Winking, J, Hecht, H.J, Schwarz, G, Mendel, R.R. | | Deposit date: | 2002-11-28 | | Release date: | 2003-12-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Active Site of the Molybdenum Cofactor Biosynthetic Protein Domain Cnx1G
Arch.Biochem.Biophys., 411, 2003
|
|
1UUY
 
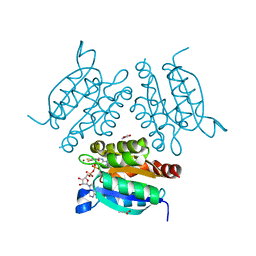 | | Structure of a molybdopterin-bound cnx1g domain links molybdenum and copper metabolism | | Descriptor: | ADENOSINE MONOPHOSPHATE, COPPER (I) ION, FORMIC ACID, ... | | Authors: | Kuper, J, Llamas, A, Hecht, H.J, Mendel, R.R, Schwarz, G. | | Deposit date: | 2004-01-12 | | Release date: | 2004-08-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of a Molybdopterin-Bound Cnx1G Domain Links Molybdenum and Copper Metabolism
Nature, 430, 2004
|
|
1UUX
 
 | | Structure of a molybdopterin-bound cnx1g domain links molybdenum and copper metabolism | | Descriptor: | COPPER (I) ION, FORMIC ACID, IMIDAZOLE, ... | | Authors: | Kuper, J, Llamas, A, Hecht, H.J, Mendel, R.R, Schwarz, G. | | Deposit date: | 2004-01-12 | | Release date: | 2004-08-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of a Molybdopterin-Bound Cnx1G Domain Links Molybdenum and Copper Metabolism
Nature, 430, 2004
|
|
1VZW
 
 | | Crystal structure of the bifunctional protein Pria | | Descriptor: | GLYCEROL, PHOSPHORIBOSYL ISOMERASE A, SULFATE ION | | Authors: | Kuper, J, Wilmanns, M. | | Deposit date: | 2004-05-27 | | Release date: | 2005-01-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Two-Fold Repeated (Beta-Alpha)(4) Half-Barrels May Provide a Molecular Tool for Dual Substrate Specificity
Embo Rep., 6, 2005
|
|
4A15
 
 | | Crystal structure of an XPD DNA complex | | Descriptor: | 5'-D(*DTP*AP*CP*GP)-3', ATP-DEPENDENT DNA HELICASE TA0057, CALCIUM ION, ... | | Authors: | Kuper, J, Wolski, S.C, Michels, G, Kisker, C. | | Deposit date: | 2011-09-14 | | Release date: | 2012-02-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional and Structural Studies of the Nucleotide Excision Repair Helicase Xpd Suggest a Polarity for DNA Translocation.
Embo J., 31, 2011
|
|
2X80
 
 | | P450 BM3 F87A in complex with DMSO | | Descriptor: | BIFUNCTIONAL P-450/NADPH-P450 REDUCTASE, DIMETHYL SULFOXIDE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Kuper, J, Wong, T.S, Roccatano, D, Wilmanns, M, Schwaneberg, U. | | Deposit date: | 2010-03-05 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Role of Active-Site Phe87 in Modulating the Organic Co-Solvent Tolerance of Cytochrome P450 Bm3 Monooxygenase.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
2X7Y
 
 | | P450 BM3 F87A in complex with DMSO | | Descriptor: | BIFUNCTIONAL P-450/NADPH-P450 REDUCTASE, DIMETHYL SULFOXIDE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Kuper, J, Wong, T.S, Roccatano, D, Wilmanns, M, Schwaneberg, U. | | Deposit date: | 2010-03-04 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Role of Active-Site Phe87 in Modulating the Organic Co-Solvent Tolerance of Cytochrome P450 Bm3 Monooxygenase.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
2J1M
 
 | | P450 BM3 Heme domain in complex with DMSO | | Descriptor: | CYTOCHROME P450 102, DIMETHYL SULFOXIDE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Kuper, J, Tuck-Seng, W, Roccatano, D, Wilmanns, M, Schwaneberg, U. | | Deposit date: | 2006-08-14 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Understanding a Mechanism of Organic Cosolvent Inactivation in Heme Monooxygenase P450 Bm-3.
J.Am.Chem.Soc., 129, 2007
|
|
2J4S
 
 | | P450 BM3 heme domain in complex with DMSO | | Descriptor: | BIFUNCTIONAL P-450:NADPH-P450 REDUCTASE, DI(HYDROXYETHYL)ETHER, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Kuper, J, Tuck-Seng, W, Roccatano, D, Wilmanns, M, Schwaneberg, U. | | Deposit date: | 2006-09-05 | | Release date: | 2007-05-15 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Understanding a Mechanism of Organic Cosolvent Inactivation in Heme Monooxygenase P450 Bm-3.
J.Am.Chem.Soc., 129, 2007
|
|
5A3D
 
 | | Structural insights into the recognition of cisplatin and AAF-dG lesions by Rad14 (XPA) | | Descriptor: | 5'-D(*DG 5IUP*GP*A 5IUP*GP*AP*CP*G 5IUP*AP*GP*AP*DGP*AP)-3', 5'-D(*DTP*CP*TP*CP*TP*AP*C 8FGP*TP*CP*AP*TP*CP*DAP*CP)-3', DNA REPAIR PROTEIN RAD14, ... | | Authors: | Kuper, J, Koch, S.C, Gasteiger, K.L, Wichlein, N, Schneider, S, Kisker, C, Carell, T. | | Deposit date: | 2015-05-28 | | Release date: | 2015-07-01 | | Last modified: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights Into the Recognition of Cisplatin and Aaf-Dg Lesion by Rad14 (Xpa).
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2VSF
 
 | | Structure of XPD from Thermoplasma acidophilum | | Descriptor: | CALCIUM ION, DNA REPAIR HELICASE RAD3 RELATED PROTEIN, IRON/SULFUR CLUSTER | | Authors: | Kuper, J, Wolski, S.C, Truglio, J.J, Kisker, C. | | Deposit date: | 2008-04-23 | | Release date: | 2008-07-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Fes Cluster-Containing Nucleotide Excision Repair Helicase Xpd.
Plos Biol., 6, 2008
|
|
2X0G
 
 | | X-RAY STRUCTURE OF A DAP-KINASE CALMODULIN COMPLEX | | Descriptor: | CALCIUM ION, CALMODULIN, DEATH-ASSOCIATED PROTEIN KINASE 1, ... | | Authors: | Kuper, J, De Diego, I, Lehmann, F, Wilmanns, M. | | Deposit date: | 2009-12-08 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular Basis of the Death-Associated Protein Kinase-Calcium/Calmodulin Regulator Complex.
Sci.Signal, 3, 2010
|
|
2Y88
 
 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS PHOSPHORIBOSYL ISOMERASE (VARIANT D11N) WITH BOUND PRFAR | | Descriptor: | PHOSPHORIBOSYL ISOMERASE A, [(2R,3S,4R,5R)-5-[4-AMINOCARBONYL-5-[[(Z)-[(3R,4R)-3,4-DIHYDROXY-2-OXO-5-PHOSPHONOOXY-PENTYL]IMINOMETHYL]AMINO]IMIDAZOL-1-YL]-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL DIHYDROGEN PHOSPHATE | | Authors: | Kuper, J, Due, A.V, Geerlof, A, Wilmanns, M. | | Deposit date: | 2011-02-03 | | Release date: | 2011-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Bisubstrate Specificity in Histidine/Tryptophan Biosynthesis Isomerase from Mycobacterium Tuberculosis by Active Site Metamorphosis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2Y85
 
 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS PHOSPHORIBOSYL ISOMERASE WITH BOUND RCDRP | | Descriptor: | 1-(O-CARBOXY-PHENYLAMINO)-1-DEOXY-D-RIBULOSE-5-PHOSPHATE, CHLORIDE ION, PHOSPHORIBOSYL ISOMERASE A, ... | | Authors: | Kuper, J, Geerlof, A, Wilmanns, M. | | Deposit date: | 2011-02-03 | | Release date: | 2011-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Bisubstrate Specificity in Histidine/Tryptophan Biosynthesis Isomerase from Mycobacterium Tuberculosis by Active Site Metamorphosis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4V98
 
 | | The 8S snRNP Assembly Intermediate | | Descriptor: | CG10419, Icln, LD23602p, ... | | Authors: | Grimm, C, Pelz, J.P, Schindelin, H, Diederichs, K, Kuper, J, Kisker, C. | | Deposit date: | 2012-05-15 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis of Assembly Chaperone- Mediated snRNP Formation.
Mol.Cell, 49, 2013
|
|
6TRS
 
 | | Crystal structure of TFIIH subunit p52 in complex with p8 | | Descriptor: | RNA polymerase II transcription factor B subunit 2, Uncharacterized protein | | Authors: | Koelmel, W, Kuper, J, Schoenwetter, E, Kisker, C. | | Deposit date: | 2019-12-19 | | Release date: | 2020-10-21 | | Last modified: | 2020-12-23 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | How to limit the speed of a motor: the intricate regulation of the XPB ATPase and translocase in TFIIH.
Nucleic Acids Res., 48, 2020
|
|
6TRU
 
 | |
2CFF
 
 | | Crystal Structure Of N-((5'-Phosphoribosyl)-Formimino)-5- Aminoimidazol-4-Carboxamid Ribonucleotid Isomerase mutant D127V (Ec 3. 1.3.15, Hisa) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-(5-PHOSPHORIBOSYL)-5-[(5-PHOSPHORIBOSYLAMINO) METHYLIDENEAMINO] IMIDAZOLE-4-CARBOXAMIDE ISOMERASE | | Authors: | He, C, Kuper, J, Sterner, R, Wilmmans, M. | | Deposit date: | 2006-02-20 | | Release date: | 2007-03-20 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of N-((5'-Phosphoribosyl)- Formimino)-5-Aminoimidazol-4-Carboxamid Ribonucleotid Isomerase Mutant D127V (Ec 3.1.3.15, Hisa)
To be Published
|
|
5A39
 
 | | Structure of Rad14 in complex with cisplatin containing DNA | | Descriptor: | 5'-D(*DGP*AP*TP*GP*AP*CP*CP*GP*TP*AP*GP*AP*GP)-3', 5'-D(*DTP*GP*AP*TP*GP*AP*CP*CP*GP*TP*AP*GP*AP)-3', Cisplatin, ... | | Authors: | Koch, S.C, Kuper, J, Gasteiger, K.L, Wichlein, N, Strasser, R, Eisen, D, Geiger, S, Schneider, S, Kisker, C, Carell, T. | | Deposit date: | 2015-05-28 | | Release date: | 2015-06-24 | | Last modified: | 2015-07-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights Into the Recognition of Cisplatin and Aaf-Dg Lesion by Rad14 (Xpa).
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4F9Z
 
 | | Crystal Structure of human ERp27 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ACETATE ION, ... | | Authors: | Kober, F.X, Koelmel, W, Kuper, J, Schindelin, H. | | Deposit date: | 2012-05-21 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of the Protein-Disulfide Isomerase Family Member ERp27 Provides Insights into Its Substrate Binding Capabilities.
J.Biol.Chem., 288, 2013
|
|
5LY9
 
 | | Structure of MITat 1.1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Variant surface glycoprotein MITAT 1.1, ... | | Authors: | Schaefer, C, Bartossek, T, Jones, N, Kuper, J, Kisker, C, Engstler, M. | | Deposit date: | 2016-09-26 | | Release date: | 2017-09-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for the shielding function of the dynamic trypanosome variant surface glycoprotein coat.
Nat Microbiol, 2, 2017
|
|
6Z4X
 
 | | Structure of the CAK complex form Chaetomium thermophilum bound to ATP-gamma-S | | Descriptor: | CHLORIDE ION, CYCLIN domain-containing protein, MAGNESIUM ION, ... | | Authors: | Peissert, S, Kuper, J, Kisker, C. | | Deposit date: | 2020-05-26 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structural basis for CDK7 activation by MAT1 and Cyclin H.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6Z3U
 
 | | Structure of the CAK complex form Chaetomium thermophilum | | Descriptor: | CHLORIDE ION, CYCLIN domain-containing protein, Protein kinase domain-containing protein, ... | | Authors: | Peissert, S, Kuper, J, Kisker, C. | | Deposit date: | 2020-05-22 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for CDK7 activation by MAT1 and Cyclin H.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
