1WNI
 
 | | Crystal Structure of H2-Proteinase | | Descriptor: | Trimerelysin II, ZINC ION | | Authors: | Kumasaka, T, Yamamoto, M, Moriyama, H, Tanaka, N, Sato, M, Katsube, Y, Yamakawa, Y, Omori-Satoh, T, Iwanaga, S, Ueki, T. | | Deposit date: | 2004-08-04 | | Release date: | 2004-08-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of H2-proteinase from the venom of Trimeresurus flavoviridis.
J.Biochem., 119, 1996
|
|
1WN5
 
 | | Crystal Structure of Blasticidin S Deaminase (BSD) Complexed with Cacodylic Acid | | Descriptor: | Blasticidin-S deaminase, CACODYLATE ION, ZINC ION | | Authors: | Kumasaka, T, Yamamoto, M, Furuichi, M, Nakasako, M, Kimura, M, Yamaguchi, I, Ueki, T. | | Deposit date: | 2004-07-27 | | Release date: | 2005-10-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of blasticidin S deaminase (BSD): implications for dynamic properties of catalytic zinc
J.Biol.Chem., 282, 2007
|
|
2CZU
 
 | | lipocalin-type prostaglandin D synthase | | Descriptor: | Prostaglandin-H2 D-isomerase | | Authors: | Kumasaka, T, Irikura, D, Ago, H, Aritake, K, Yamamoto, M, Inoue, T, Miyano, M, Urade, Y, Hayaishi, O, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-17 | | Release date: | 2006-10-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of the catalytic mechanism operating in open-closed conformers of lipocalin type prostaglandin D synthase.
J.Biol.Chem., 284, 2009
|
|
2CZT
 
 | | lipocalin-type prostaglandin D synthase | | Descriptor: | Prostaglandin-H2 D-isomerase | | Authors: | Kumasaka, T, Irikura, D, Ago, H, Aritake, K, Yamamoto, M, Inoue, T, Miyano, M, Urade, Y, Hayaishi, O, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-17 | | Release date: | 2006-10-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the catalytic mechanism operating in open-closed conformers of lipocalin type prostaglandin D synthase.
J.Biol.Chem., 284, 2009
|
|
1WN6
 
 | | Crystal Structure of Blasticidin S Deaminase (BSD) Complexed with Tetrahedral Intermediate of Blasticidin S | | Descriptor: | 6-(4-AMINO-4-HYDROXY-2-OXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-3-[3-AMINO-5-(N-METHYL-GUANIDINO)-PENT ANOYLAMINO]-3,6-DIHYDRO-2H-PYRAN-2-CARBOXYLIC ACID, ARSENIC, Blasticidin-S deaminase, ... | | Authors: | Kumasaka, T, Yamamoto, M, Furuichi, M, Nakasako, M, Kimura, M, Yamaguchi, I, Ueki, T. | | Deposit date: | 2004-07-27 | | Release date: | 2005-10-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of blasticidin S deaminase (BSD): implications for dynamic properties of catalytic zinc
J.Biol.Chem., 282, 2007
|
|
2Z3H
 
 | | Crystal structure of blasticidin S deaminase (BSD) complexed with deaminohydroxy blasticidin S | | Descriptor: | 1-(4-{[(3R)-3-AMINO-5-{[(Z)-AMINO(IMINO)METHYL](METHYL)AMINO}PENTANOYL]AMINO}-2,3,4-TRIDEOXY-D-ERYTHRO-HEX-2-ENOPYRANURONOSYL)-4-HYDROXYPYRIMIDIN-2(1H)-ONE, Blasticidin-S deaminase, ZINC ION | | Authors: | Kumasaka, T, Yamamoto, M, Furuichi, M, Nakasako, M, Kimura, M, Yamaguchi, I, Ueki, T. | | Deposit date: | 2007-06-04 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of blasticidin S deaminase (BSD): implications for dynamic properties of catalytic zinc
J.Biol.Chem., 282, 2007
|
|
2Z3G
 
 | | Crystal structure of blasticidin S deaminase (BSD) | | Descriptor: | Blasticidin-S deaminase, ZINC ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Kumasaka, T, Yamamoto, M, Furuichi, M, Nakasako, M, Kimura, M, Yamaguchi, I, Ueki, T. | | Deposit date: | 2007-06-04 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of blasticidin S deaminase (BSD): implications for dynamic properties of catalytic zinc
J.Biol.Chem., 282, 2007
|
|
2Z3I
 
 | | Crystal structure of blasticidin S deaminase (BSD) mutant E56Q complexed with substrate | | Descriptor: | BLASTICIDIN S, Blasticidin-S deaminase, CACODYLATE ION, ... | | Authors: | Kumasaka, T, Yamamoto, M, Furuichi, M, Nakasako, M, Kimura, M, Yamaguchi, I, Ueki, T. | | Deposit date: | 2007-06-04 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of blasticidin S deaminase (BSD): implications for dynamic properties of catalytic zinc
J.Biol.Chem., 282, 2007
|
|
8GOH
 
 | |
8GOI
 
 | |
5B2Z
 
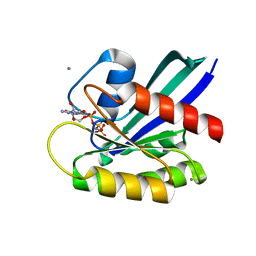 | | H-Ras WT in complex with GppNHp (state 2*) before structural transition by humidity control | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Kumasaka, T, Miyano, N, Baba, S, Matsumoto, S, Kataoka, T, Shima, F. | | Deposit date: | 2016-02-07 | | Release date: | 2016-06-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Molecular Mechanism for Conformational Dynamics of Ras-GTP Elucidated from In-Situ Structural Transition in Crystal
Sci Rep, 6, 2016
|
|
8GOJ
 
 | |
5B30
 
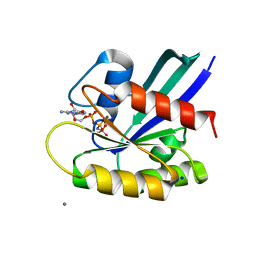 | | H-Ras WT in complex with GppNHp (state 1) after structural transition by humidity control | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Kumasaka, T, Miyano, N, Baba, S, Matsumoto, S, Kataoka, T, Shima, F. | | Deposit date: | 2016-02-08 | | Release date: | 2016-06-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular Mechanism for Conformational Dynamics of Ras-GTP Elucidated from In-Situ Structural Transition in Crystal
Sci Rep, 6, 2016
|
|
5B4S
 
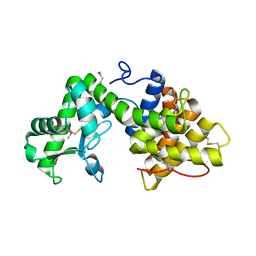 | | Crystal Structure of GH80 chitosanase from Mitsuaria chitosanitabida | | Descriptor: | Chitosanase | | Authors: | Kumasaka, T, Yorinaga, Y, Yamamoto, M, Hamada, K, Kawamukai, M. | | Deposit date: | 2016-04-19 | | Release date: | 2017-02-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a family 80 chitosanase from Mitsuaria chitosanitabida
FEBS Lett., 591, 2017
|
|
3AH7
 
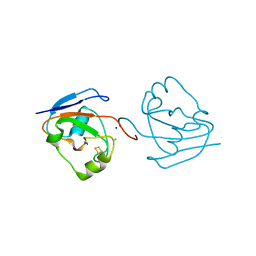 | | Crystal structure of the ISC-like [2Fe-2S] ferredoxin (FdxB) from Pseudomonas putida JCM 20004 | | Descriptor: | CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, SODIUM ION, ... | | Authors: | Kumasaka, T, Shimizu, N, Ohmori, D, Iwasaki, T. | | Deposit date: | 2010-04-20 | | Release date: | 2011-04-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the ISC-like [2Fe-2S] ferredoxin (FdxB) from Pseudomonas putida JCM 20004
To be Published
|
|
2Z3J
 
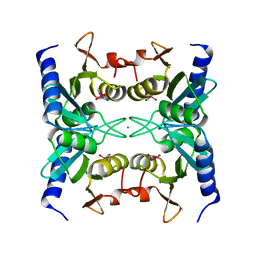 | | Crystal structure of blasticidin S deaminase (BSD) R90K mutant | | Descriptor: | Blasticidin-S deaminase, CACODYLATE ION, CHLORIDE ION, ... | | Authors: | Teh, A.H, Kumasaka, T, Yamamoto, M, Furuichi, M, Nakasako, M, Kimura, M, Yamaguchi, I, Ueki, T. | | Deposit date: | 2007-06-04 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of blasticidin S deaminase (BSD): implications for dynamic properties of catalytic zinc
J.Biol.Chem., 282, 2007
|
|
3HSB
 
 | | Crystal structure of YmaH (Hfq) from Bacillus subtilis in complex with an RNA aptamer | | Descriptor: | Protein hfq, RNA (5'-R(*AP*GP*AP*GP*AP*GP*A)-3') | | Authors: | Baba, S, Someya, T, Kumasaka, T, Kawai, G, Nakamura, K. | | Deposit date: | 2009-06-10 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Hfq from Bacillus subtilis in complex with SELEX-derived RNA aptamer: insight into RNA-binding properties of bacterial Hfq
Nucleic Acids Res., 40, 2012
|
|
6J8M
 
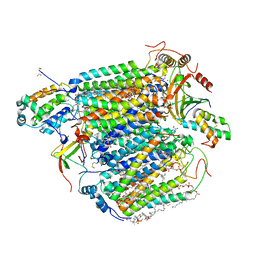 | | Low-dose structure of bovine heart cytochrome c oxidase in the fully oxidized state determined using 30 keV X-ray | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Ueno, G, Shimada, A, Yamashita, E, Hasegawa, K, Kumasaka, T, Shinzawa-Itoh, K, Yoshikawa, S, Tsukihara, T, Yamamoto, M. | | Deposit date: | 2019-01-20 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Low-dose X-ray structure analysis of cytochrome c oxidase utilizing high-energy X-rays.
J.Synchrotron Radiat., 26, 2019
|
|
2HYK
 
 | | The crystal structure of an endo-beta-1,3-glucanase from alkaliphilic Nocardiopsis sp.strain F96 | | Descriptor: | Beta-1,3-glucanase, CALCIUM ION, ETHANOL, ... | | Authors: | Fibriansah, G, Nakamura, S, Kumasaka, T. | | Deposit date: | 2006-08-07 | | Release date: | 2007-10-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The 1.3 A crystal structure of a novel endo-beta-1,3-glucanase of glycoside hydrolase family 16 from alkaliphilic Nocardiopsis sp. strain F96.
Proteins, 69, 2007
|
|
1EE8
 
 | | CRYSTAL STRUCTURE OF MUTM (FPG) PROTEIN FROM THERMUS THERMOPHILUS HB8 | | Descriptor: | MUTM (FPG) PROTEIN, ZINC ION | | Authors: | Sugahara, M, Mikawa, T, Kumasaka, T, Yamamoto, M, Kato, R, Fukuyama, K, Inoue, Y, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2000-01-31 | | Release date: | 2001-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a repair enzyme of oxidatively damaged DNA, MutM (Fpg), from an extreme thermophile, Thermus thermophilus HB8.
EMBO J., 19, 2000
|
|
3CE1
 
 | | Crystal Structure of the Cu/Zn Superoxide Dismutase from Cryptococcus liquefaciens Strain N6 | | Descriptor: | ACETATE ION, COPPER (II) ION, Superoxide dismutase [Cu-Zn], ... | | Authors: | Teh, A.H, Kanamasa, S, Kajiwara, S, Kumasaka, T. | | Deposit date: | 2008-02-27 | | Release date: | 2008-08-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of Cu/Zn superoxide dismutase from the heavy-metal-tolerant yeast Cryptococcus liquefaciens strain N6.
Biochem.Biophys.Res.Commun., 374, 2008
|
|
1WQS
 
 | | Crystal structure of Norovirus 3C-like protease | | Descriptor: | 3C-like protease, D(-)-TARTARIC ACID, L(+)-TARTARIC ACID, ... | | Authors: | Nakamura, K, Someya, Y, Kumasaka, T, Tanaka, N. | | Deposit date: | 2004-10-01 | | Release date: | 2005-10-04 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A norovirus protease structure provides insights into active and substrate binding site integrity
J.Virol., 79, 2005
|
|
1WPR
 
 | | Crystal structure of RsbQ inhibited by PMSF | | Descriptor: | GLYCEROL, Sigma factor sigB regulation protein rsbQ, phenylmethanesulfonic acid | | Authors: | Kaneko, T, Tanaka, N, Kumasaka, T. | | Deposit date: | 2004-09-11 | | Release date: | 2005-02-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of RsbQ, a stress-response regulator in Bacillus subtilis
Protein Sci., 14, 2005
|
|
8YOZ
 
 | | Crystal structure of the small zinc-finger protein ZifS (TTHA0897) from Thermus thermophilus HB8. | | Descriptor: | Transcription factor zinc-finger domain-containing protein, ZINC ION | | Authors: | Kurinami, S, Fukui, K, Murakawa, T, Baba, S, Kumasaka, T, Okanishi, H, Kanai, Y, Yano, T, Masui, R. | | Deposit date: | 2024-03-14 | | Release date: | 2025-03-19 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | The crystal structure of the small zinc-finger protein ZifS from Thermus thermophilus HB8.
J.Biochem., 2025
|
|
2GRU
 
 | | Crystal structure of 2-deoxy-scyllo-inosose synthase complexed with carbaglucose-6-phosphate, NAD+ and Co2+ | | Descriptor: | (1R,2S,3S,4R)-5-METHYLENECYCLOHEXANE-1,2,3,4-TETRAOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-deoxy-scyllo-inosose synthase, ... | | Authors: | Nango, E, Kumasaka, T. | | Deposit date: | 2006-04-25 | | Release date: | 2007-05-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of 2-deoxy-scyllo-inosose synthase, a key enzyme in the biosynthesis of 2-deoxystreptamine-containing aminoglycoside antibiotics, in complex with a mechanism-based inhibitor and NAD+
Proteins, 70, 2008
|
|
