2Z9Z
 
 | | Crystal structure of CERT START domain(N504A mutant), in complex with C10-diacylglycerol | | Descriptor: | (2S)-3-hydroxypropane-1,2-diyl didecanoate, Lipid-transfer protein CERT | | Authors: | Kudo, N, Kumagai, K, Wakatsuki, S, Nishijima, M, Hanada, K, Kato, R. | | Deposit date: | 2007-09-26 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural basis for specific lipid recognition by CERT responsible for nonvesicular trafficking of ceramide.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2Z9Y
 
 | | Crystal structure of CERT START domain in complex with C10-diacylglycerol | | Descriptor: | (2S)-3-hydroxypropane-1,2-diyl didecanoate, Lipid-transfer protein CERT | | Authors: | Kudo, N, Kumagai, K, Wakatsuki, S, Nishijima, M, Hanada, K, Kato, R. | | Deposit date: | 2007-09-26 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for specific lipid recognition by CERT responsible for nonvesicular trafficking of ceramide.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
5Y0Z
 
 | | Human SIRT2 in complex with a specific inhibitor, NPD11033 | | Descriptor: | (1~{R},9~{S})-11-[(2~{R})-3-[2,4-bis(2-methylbutan-2-yl)phenoxy]-2-oxidanyl-propyl]-7,11-diazatricyclo[7.3.1.0^{2,7}]trideca-2,4-dien-6-one, NAD-dependent protein deacetylase sirtuin-2, ZINC ION | | Authors: | Kudo, N, Ito, A, Yoshida, M. | | Deposit date: | 2017-07-19 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of a novel small molecule that inhibits deacetylase but not defatty-acylase reaction catalysed by SIRT2.
Philos. Trans. R. Soc. Lond., B, Biol. Sci., 373, 2018
|
|
3H3S
 
 | | Crystal structure of the CERT START domain in complex with HPA-15 | | Descriptor: | Goodpasture antigen binding protein, N-[(1R,3R)-3-hydroxy-1-(hydroxymethyl)-3-phenylpropyl]pentadecanamide | | Authors: | Kudo, N, Wakatsuki, S, Kato, R. | | Deposit date: | 2009-04-17 | | Release date: | 2010-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structures of the CERT START domain with inhibitors provide insights into the mechanism of ceramide transfer.
J.Mol.Biol., 396, 2010
|
|
3H3T
 
 | | Crystal structure of the CERT START domain in complex with HPA-16 | | Descriptor: | Goodpasture antigen binding protein, N-[(1R,3R)-3-hydroxy-1-(hydroxymethyl)-3-phenylpropyl]hexadecanamide | | Authors: | Kudo, N, Wakatsuki, S, Kato, R. | | Deposit date: | 2009-04-17 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the CERT START domain with inhibitors provide insights into the mechanism of ceramide transfer.
J.Mol.Biol., 396, 2010
|
|
3H3Q
 
 | | Crystal structure of the CERT START domain in complex with HPA-13 | | Descriptor: | Goodpasture antigen binding protein, N-[(1R,3R)-3-hydroxy-1-(hydroxymethyl)-3-phenylpropyl]tridecanamide | | Authors: | Kudo, N, Wakatsuki, S, Kato, R. | | Deposit date: | 2009-04-17 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the CERT START domain with inhibitors provide insights into the mechanism of ceramide transfer.
J.Mol.Biol., 396, 2010
|
|
3H3R
 
 | | Crystal structure of the CERT START domain in complex with HPA-14 | | Descriptor: | Goodpasture antigen binding protein, N-[(1R,3R)-3-hydroxy-1-(hydroxymethyl)-3-phenylpropyl]tetradecanamide | | Authors: | Kudo, N, Wakatsuki, S, Kato, R. | | Deposit date: | 2009-04-17 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of the CERT START domain with inhibitors provide insights into the mechanism of ceramide transfer.
J.Mol.Biol., 396, 2010
|
|
2E3Q
 
 | | Crystal structure of CERT START domain in complex with C18-ceramide (P212121) | | Descriptor: | DIMETHYL SULFOXIDE, Lipid-transfer protein CERT, N-((E,2S,3R)-1,3-DIHYDROXYOCTADEC-4-EN-2-YL)STEARAMIDE | | Authors: | Kudo, N, Kumagai, K, Wakatsuki, S, Nishijima, M, Hanada, K, Kato, R. | | Deposit date: | 2006-11-28 | | Release date: | 2007-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural basis for specific lipid recognition by CERT responsible for nonvesicular trafficking of ceramide.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2E3P
 
 | | Crystal structure of CERT START domain in complex with C16-cearmide (P1) | | Descriptor: | Lipid-transfer protein CERT, N-((E,2S,3R)-1,3-DIHYDROXYOCTADEC-4-EN-2-YL)PALMITAMIDE | | Authors: | Kudo, N, Kumagai, K, Wakatsuki, S, Nishijima, M, Hanada, K, Kato, R. | | Deposit date: | 2006-11-28 | | Release date: | 2007-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for specific lipid recognition by CERT responsible for nonvesicular trafficking of ceramide.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2E3M
 
 | | Crystal structure of CERT START domain | | Descriptor: | Lipid-transfer protein CERT | | Authors: | Kudo, N, Kumagai, K, Wakatsuki, S, Nishijima, M, Hanada, K, Kato, R. | | Deposit date: | 2006-11-28 | | Release date: | 2007-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for specific lipid recognition by CERT responsible for nonvesicular trafficking of ceramide.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2E3N
 
 | | Crystal structure of CERT START domain in complex with C6-ceramide (P212121) | | Descriptor: | Lipid-transfer protein CERT, N-((E,2S,3R)-1,3-DIHYDROXYOCTADEC-4-EN-2-YL)HEXANAMIDE | | Authors: | Kudo, N, Kumagai, K, Wakatsuki, S, Nishijima, M, Hanada, K, Kato, R. | | Deposit date: | 2006-11-28 | | Release date: | 2007-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for specific lipid recognition by CERT responsible for nonvesicular trafficking of ceramide.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2E3R
 
 | | Crystal structure of CERT START domain in complex with C18-ceramide (P1) | | Descriptor: | Lipid-transfer protein CERT, N-((E,2S,3R)-1,3-DIHYDROXYOCTADEC-4-EN-2-YL)STEARAMIDE | | Authors: | Kudo, N, Kumagai, K, Wakatsuki, S, Nishijima, M, Hanada, K, Kato, R. | | Deposit date: | 2006-11-28 | | Release date: | 2007-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for specific lipid recognition by CERT responsible for nonvesicular trafficking of ceramide.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2E3S
 
 | | Crystal structure of CERT START domain co-crystallized with C24-ceramide (P21) | | Descriptor: | Lipid-transfer protein CERT | | Authors: | Kudo, N, Kumagai, K, Wakatsuki, S, Nishijima, M, Hanada, K, Kato, R. | | Deposit date: | 2006-11-28 | | Release date: | 2007-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural basis for specific lipid recognition by CERT responsible for nonvesicular trafficking of ceramide.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2E3O
 
 | | Crystal structure of CERT START domain in complex with C16-ceramide (P212121) | | Descriptor: | DIMETHYL SULFOXIDE, Lipid-transfer protein CERT, N-((E,2S,3R)-1,3-DIHYDROXYOCTADEC-4-EN-2-YL)PALMITAMIDE | | Authors: | Kudo, N, Kumagai, K, Wakatsuki, S, Nishijima, M, Hanada, K, Kato, R. | | Deposit date: | 2006-11-28 | | Release date: | 2007-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for specific lipid recognition by CERT responsible for nonvesicular trafficking of ceramide.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3VTG
 
 | | High choriolytic enzyme 1 (HCE-1), a hatching enzyme zinc-protease from Oryzias latipes (Medaka fish) | | Descriptor: | High choriolytic enzyme 1, ZINC ION | | Authors: | Kudo, N, Yasumasu, S, Iuchi, I, Tanokura, M. | | Deposit date: | 2012-05-30 | | Release date: | 2013-06-05 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal Structure of High choriolytic enzyme 1 (HCE-1), a hatching enzyme from Oryzias latipes (Medaka fish)
To be Published
|
|
3VVH
 
 | |
5Y4H
 
 | | Human SIRT3 in complex with halistanol sulfate | | Descriptor: | NAD-dependent protein deacetylase sirtuin-3, mitochondrial, THr-Arg-Ser-GLY-ALY-VAL-MET-ARG-ARG-LEU-ARG-ARG, ... | | Authors: | Kudo, N, Ito, A, Yoshida, M. | | Deposit date: | 2017-08-03 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Halistanol sulfates I and J, new SIRT1-3 inhibitory steroid sulfates from a marine sponge of the genus Halichondria
J. Antibiot., 71, 2018
|
|
7BOT
 
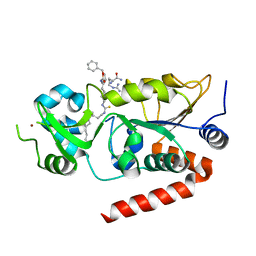 | | Human SIRT2 in complex with myristoyl thiourea inhibitor, No.23 | | Descriptor: | N-dodecylmethanethioamide, NAD-dependent protein deacetylase sirtuin-2, ZINC ION, ... | | Authors: | Kudo, N, Olsen, C.A, Minoru, Y. | | Deposit date: | 2020-03-19 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism-based inhibitors of SIRT2: structure-activity relationship, X-ray structures, target engagement, regulation of alpha-tubulin acetylation and inhibition of breast cancer cell migration.
Rsc Chem Biol, 2, 2021
|
|
7BOS
 
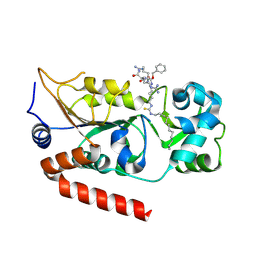 | | Human SIRT2 in complex with myristoyl thiourea inhibitor, No.13 | | Descriptor: | Myristoyl thiourea inhibitor, No.13, N-dodecylmethanethioamide, ... | | Authors: | Kudo, N, Olsen, C.A, Minoru, Y. | | Deposit date: | 2020-03-19 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism-based inhibitors of SIRT2: structure-activity relationship, X-ray structures, target engagement, regulation of alpha-tubulin acetylation and inhibition of breast cancer cell migration.
Rsc Chem Biol, 2, 2021
|
|
4Y6L
 
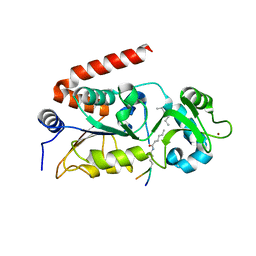 | | Human SIRT2 in complex with myristoylated peptide (H3K9myr) | | Descriptor: | NAD-dependent protein deacetylase sirtuin-2, ZINC ION, peptide THR-ALA-ARG-MYK-SER-THR-GLY | | Authors: | Kudo, N, Ito, A, Yoshida, M. | | Deposit date: | 2015-02-13 | | Release date: | 2016-01-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Kinetic and Structural Basis for Acyl-Group Selectivity and NAD(+) Dependence in Sirtuin-Catalyzed Deacylation.
Biochemistry, 54, 2015
|
|
4Y6Q
 
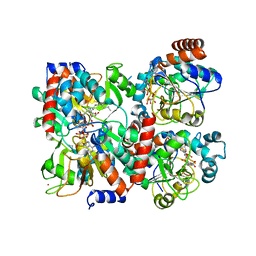 | | Human SIRT2 in complex with 2-O-myristoyl-ADP-ribose | | Descriptor: | NAD-dependent protein deacetylase sirtuin-2, ZINC ION, [(2S,3R,4R,5R)-5-[[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxymethyl]-2,4-bis(oxidanyl)oxolan-3-yl] tetradecanoate | | Authors: | Kudo, N, Ito, A, Yoshida, M. | | Deposit date: | 2015-02-13 | | Release date: | 2016-01-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Kinetic and Structural Basis for Acyl-Group Selectivity and NAD(+) Dependence in Sirtuin-Catalyzed Deacylation.
Biochemistry, 54, 2015
|
|
4Y6O
 
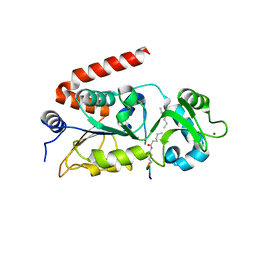 | |
3AAD
 
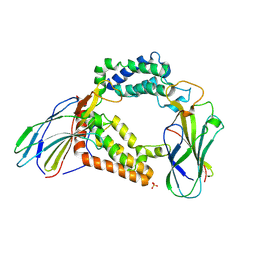 | | Structure of the histone chaperone CIA/ASF1-double bromodomain complex linking histone modifications and site-specific histone eviction | | Descriptor: | Histone chaperone ASF1A, SULFATE ION, Transcription initiation factor TFIID subunit 1 | | Authors: | Akai, Y, Adachi, N, Hayashi, Y, Eitoku, M, Sano, N, Natsume, R, Kudo, N, Tanokura, M, Senda, T, Horikoshi, M. | | Deposit date: | 2009-11-16 | | Release date: | 2010-04-28 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the histone chaperone CIA/ASF1-double bromodomain complex linking histone modifications and site-specific histone eviction
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1V3Y
 
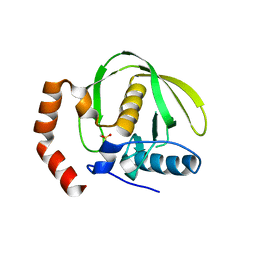 | | The crystal structure of peptide deformylase from Thermus thermophilus HB8 | | Descriptor: | Peptide deformylase | | Authors: | Kamo, M, Kudo, N, Lee, W.C, Ito, K, Motoshim, H, Tanokura, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-07 | | Release date: | 2004-12-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The crystal structure of peptide deformylase from Thermus thermophilus HB8
to be published
|
|
1EQK
 
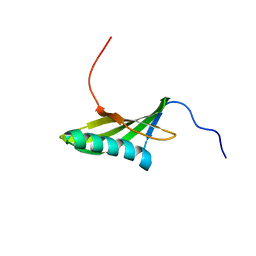 | | SOLUTION STRUCTURE OF ORYZACYSTATIN-I, A CYSTEINE PROTEINASE INHIBITOR OF THE RICE, ORYZA SATIVA L. JAPONICA | | Descriptor: | ORYZACYSTATIN-I | | Authors: | Nagata, K, Kudo, N, Abe, K, Arai, S, Tanokura, M. | | Deposit date: | 2000-04-05 | | Release date: | 2001-01-10 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of oryzacystatin-I, a cysteine proteinase inhibitor of the rice, Oryza sativa L. japonica.
Biochemistry, 39, 2000
|
|
