2X7O
 
 | | Crystal structure of TGFbRI complexed with an indolinone inhibitor | | Descriptor: | (3Z)-N-ETHYL-N-METHYL-2-OXO-3-(PHENYL{[4-(PIPERIDIN-1-YLMETHYL)PHENYL]AMINO}METHYLIDENE)-2,3-DIHYDRO-1H-INDOLE-6-CARBOXAMIDE, TGF-BETA RECEPTOR TYPE I | | Authors: | Roth, G.J, Heckel, A, Brandl, T, Grauert, M, Hoerer, S, Kley, J.T, Schnapp, G, Baum, P, Mennerich, D, Schnapp, A, Park, J.E. | | Deposit date: | 2010-03-03 | | Release date: | 2010-10-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Design, Synthesis and Evaluation of Indolinones as Inhibitors of the Transforming Growth Factor Beta Receptor I (Tgfbri)
J.Med.Chem., 53, 2010
|
|
1PRE
 
 | | PROAEROLYSIN | | Descriptor: | PROAEROLYSIN | | Authors: | Parker, M.W, Buckley, J.T, Postma, J.P.M, Tucker, A.D, Tsernoglou, D. | | Deposit date: | 1995-09-15 | | Release date: | 1996-10-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Aeromonas toxin proaerolysin in its water-soluble and membrane-channel states.
Nature, 367, 1994
|
|
9FHE
 
 | | hKHK-C in complex with BI-9787 (pH 5.5) | | Descriptor: | (2~{S})-3-[3-[[4-[bis(fluoranyl)methyl]-3-cyano-6-[(3~{S})-3-(dimethylamino)pyrrolidin-1-yl]pyridin-2-yl]amino]-4-methylsulfanyl-phenyl]-2-methyl-propanoic acid, Ketohexokinase | | Authors: | Ebenhoch, R, Pautsch, A. | | Deposit date: | 2024-05-27 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.313 Å) | | Cite: | Discovery of BI-9787, a potent zwitterionic ketohexokinase inhibitor with oral bioavailability.
Bioorg.Med.Chem.Lett., 112, 2024
|
|
9FHD
 
 | | hKHK-C in fomplex with BI-9787 | | Descriptor: | (2~{S})-3-[3-[[4-[bis(fluoranyl)methyl]-3-cyano-6-[(3~{S})-3-(dimethylamino)pyrrolidin-1-yl]pyridin-2-yl]amino]-4-methylsulfanyl-phenyl]-2-methyl-propanoic acid, Ketohexokinase, SULFATE ION | | Authors: | Ebenhoch, R, Pautsch, A. | | Deposit date: | 2024-05-27 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | Discovery of BI-9787, a potent zwitterionic ketohexokinase inhibitor with oral bioavailability.
Bioorg.Med.Chem.Lett., 112, 2024
|
|
8OME
 
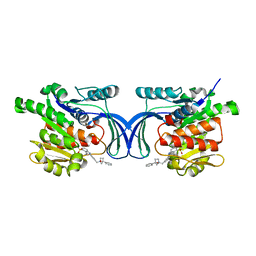 | | Crystal structure of hKHK-A in complex with compound-4 | | Descriptor: | Ketohexokinase, compound | | Authors: | Ebenhoch, R, Pautsch, A. | | Deposit date: | 2023-03-31 | | Release date: | 2023-09-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of human and mouse ketohexokinase provide a structural basis for species- and isoform-selective inhibitor design.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8OMD
 
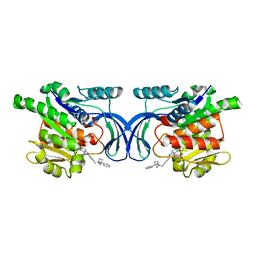 | | Crystal structure of mKHK in complex with compound-4 | | Descriptor: | Ketohexokinase, compound | | Authors: | Ebenhoch, R, Pautsch, A. | | Deposit date: | 2023-03-31 | | Release date: | 2023-09-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of human and mouse ketohexokinase provide a structural basis for species- and isoform-selective inhibitor design.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8OMF
 
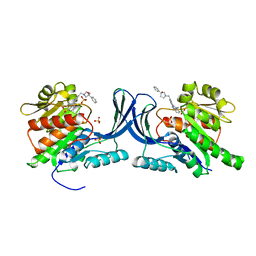 | | Crystal structure of hKHK-C in complex with compound-4 | | Descriptor: | Ketohexokinase, SULFATE ION, compound | | Authors: | Ebenhoch, R, Pautsch, A. | | Deposit date: | 2023-03-31 | | Release date: | 2023-09-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structures of human and mouse ketohexokinase provide a structural basis for species- and isoform-selective inhibitor design.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8OMG
 
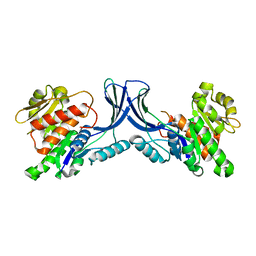 | | Crystal structure of mKHK (apo) | | Descriptor: | Ketohexokinase | | Authors: | Ebenhoch, E, Pautsch, P. | | Deposit date: | 2023-03-31 | | Release date: | 2023-09-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structures of human and mouse ketohexokinase provide a structural basis for species- and isoform-selective inhibitor design.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8OMJ
 
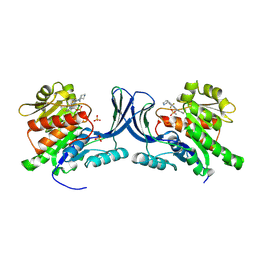 | | hKHK-C in complex with compound 28 | | Descriptor: | Ketohexokinase, SULFATE ION, [3-[[6-[(3~{a}~{R},6~{a}~{S})-2,3,3~{a},4,6,6~{a}-hexahydro-1~{H}-pyrrolo[3,4-c]pyrrol-5-yl]-3-cyano-4-(trifluoromethyl)pyridin-2-yl]amino]-4-methylsulfanyl-phenyl]methoxy-methyl-phosphinic acid | | Authors: | Pautsch, A, Ebenhoch, R. | | Deposit date: | 2023-03-31 | | Release date: | 2024-07-10 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.978 Å) | | Cite: | Discovery of BI-9787, a potent zwitterionic ketohexokinase inhibitor with oral bioavailability.
Bioorg.Med.Chem.Lett., 112, 2024
|
|
8OMK
 
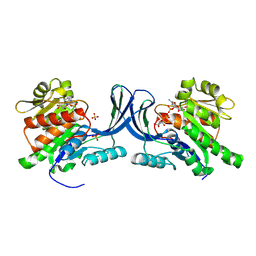 | | hKHK-C in complex with ADP & fructose 1-phosphate | | Descriptor: | 1-O-phosphono-beta-D-fructofuranose, ADENOSINE-5'-DIPHOSPHATE, Ketohexokinase, ... | | Authors: | Ebenhoch, R, Pautsch, A. | | Deposit date: | 2023-03-31 | | Release date: | 2024-01-10 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Discovery of BI-9787, a potent zwitterionic ketohexokinase inhibitor with oral bioavailability.
Bioorg.Med.Chem.Lett., 112, 2024
|
|
3C0N
 
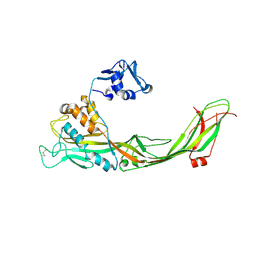 | | Crystal structure of the proaerolysin mutant Y221G at 2.2 A | | Descriptor: | Aerolysin | | Authors: | Pernot, L, Schiltz, M, Thurnheer, S, Burr, S.E, van der Goot, G. | | Deposit date: | 2008-01-21 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular assembly of the aerolysin pore reveals a swirling membrane-insertion mechanism.
Nat.Chem.Biol., 9, 2013
|
|
3C0O
 
 | | Crystal structure of the proaerolysin mutant Y221G complexed with mannose-6-phosphate | | Descriptor: | 6-O-phosphono-alpha-D-mannopyranose, ACETATE ION, Aerolysin | | Authors: | Pernot, L, Schiltz, M, Thurnheer, S, Burr, S.E, van der Goot, G. | | Deposit date: | 2008-01-21 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular assembly of the aerolysin pore reveals a swirling membrane-insertion mechanism.
Nat.Chem.Biol., 9, 2013
|
|
3C0M
 
 | | Crystal structure of the proaerolysin mutant Y221G | | Descriptor: | Aerolysin | | Authors: | Pernot, L, Schiltz, M, Thurnheer, S, Burr, S.E, van der Goot, G. | | Deposit date: | 2008-01-21 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Molecular assembly of the aerolysin pore reveals a swirling membrane-insertion mechanism.
Nat.Chem.Biol., 9, 2013
|
|
3G4O
 
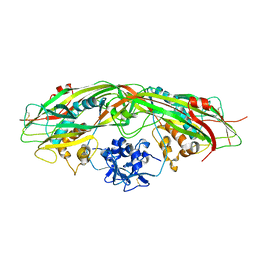 | |
3G4N
 
 | |
