2YYL
 
 | | Crystal structure of the mutant of HpaB (T198I, A276G, and R466H) complexed with FAD | | Descriptor: | 4-hydroxyphenylacetate-3-hydroxylase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Kim, S.-H, Hisano, T, Takeda, K, Iwasaki, W, Ebihara, A, Miki, K. | | Deposit date: | 2007-04-30 | | Release date: | 2007-09-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the Oxygenase Component (HpaB) of the 4-Hydroxyphenylacetate 3-Monooxygenase from Thermus thermophilus HB8
J.Biol.Chem., 282, 2007
|
|
2YYI
 
 | | Crystal structure of the oxygenase component (HpaB) of 4-hydroxyphenylacetate 3-monooxygenase complexed with FAD | | Descriptor: | 4-hydroxyphenylacetate-3-hydroxylase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Kim, S.-H, Hisano, T, Takeda, K, Iwasaki, W, Ebihara, A, Miki, K. | | Deposit date: | 2007-04-30 | | Release date: | 2007-09-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal Structure of the Oxygenase Component (HpaB) of the 4-Hydroxyphenylacetate 3-Monooxygenase from Thermus thermophilus HB8
J.Biol.Chem., 282, 2007
|
|
2YYG
 
 | | Crystal structure of the oxygenase component (HpaB) of 4-hydroxyphenylacetate 3-monooxygenase | | Descriptor: | 4-hydroxyphenylacetate-3-hydroxylase, SULFATE ION | | Authors: | Kim, S.-H, Hisano, T, Takeda, K, Iwasaki, W, Ebihara, A, Miki, K. | | Deposit date: | 2007-04-30 | | Release date: | 2007-09-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Oxygenase Component (HpaB) of the 4-Hydroxyphenylacetate 3-Monooxygenase from Thermus thermophilus HB8
J.Biol.Chem., 282, 2007
|
|
2YYJ
 
 | | Crystal structure of the oxygenase component (HpaB) of 4-hydroxyphenylacetate 3-monooxygenase complexed with FAD and 4-hydroxyphenylacetate | | Descriptor: | 4-HYDROXYPHENYLACETATE, 4-hydroxyphenylacetate-3-hydroxylase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kim, S.-H, Hisano, T, Takeda, K, Iwasaki, W, Ebihara, A, Miki, K. | | Deposit date: | 2007-04-30 | | Release date: | 2007-09-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal Structure of the Oxygenase Component (HpaB) of the 4-Hydroxyphenylacetate 3-Monooxygenase from Thermus thermophilus HB8
J.Biol.Chem., 282, 2007
|
|
2YYM
 
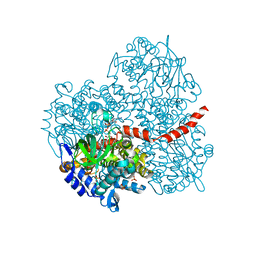 | | Crystal structure of the mutant of HpaB (T198I, A276G, and R466H) complexed with FAD and 4-hydroxyphenylacetate | | Descriptor: | 4-HYDROXYPHENYLACETATE, 4-hydroxyphenylacetate-3-hydroxylase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kim, S.-H, Hisano, T, Takeda, K, Iwasaki, W, Ebihara, A, Miki, K. | | Deposit date: | 2007-04-30 | | Release date: | 2007-09-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the Oxygenase Component (HpaB) of the 4-Hydroxyphenylacetate 3-Monooxygenase from Thermus thermophilus HB8
J.Biol.Chem., 282, 2007
|
|
2YYK
 
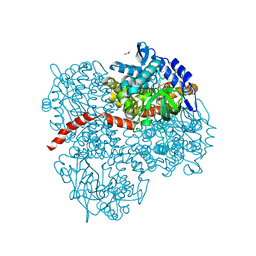 | | Crystal structure of the mutant of HpaB (T198I, A276G, and R466H) | | Descriptor: | 4-hydroxyphenylacetate-3-hydroxylase, ACETIC ACID, GLYCEROL, ... | | Authors: | Kim, S.-H, Hisano, T, Takeda, K, Iwasaki, W, Ebihara, A, Miki, K. | | Deposit date: | 2007-04-30 | | Release date: | 2007-09-04 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Oxygenase Component (HpaB) of the 4-Hydroxyphenylacetate 3-Monooxygenase from Thermus thermophilus HB8
J.Biol.Chem., 282, 2007
|
|
1LAH
 
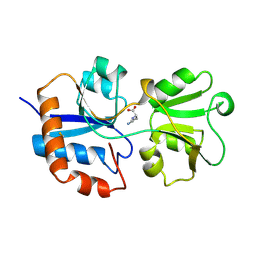 | | STRUCTURAL BASES FOR MULTIPLE LIGAND SPECIFICITY OF THE PERIPLASMIC LYSINE-, ARGININE-, ORNITHINE-BINDING PROTEIN | | Descriptor: | L-ornithine, LYSINE, ARGININE, ... | | Authors: | Kim, S.-H, Oh, B.-H. | | Deposit date: | 1993-10-06 | | Release date: | 1995-07-10 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis for multiple ligand specificity of the periplasmic lysine-, arginine-, ornithine-binding protein.
J.Biol.Chem., 269, 1994
|
|
6Q21
 
 | |
4Q21
 
 | |
2Q21
 
 | |
2LIG
 
 | | THREE-DIMENSIONAL STRUCTURES OF THE LIGAND-BINDING DOMAIN OF THE BACTERIAL ASPARTATE RECEPTOR WITH AND WITHOUT A LIGAND | | Descriptor: | 1,10-PHENANTHROLINE, ASPARTATE RECEPTOR, ASPARTIC ACID, ... | | Authors: | Kim, S.-H, Yeh, J.I, Prive, G.G, Milburn, M, Scott, W, Koshland Junior, D.E. | | Deposit date: | 1995-04-18 | | Release date: | 1995-09-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand.
Science, 254, 1991
|
|
1LIH
 
 | | THREE-DIMENSIONAL STRUCTURES OF THE LIGAND-BINDING DOMAIN OF THE BACTERIAL ASPARTATE RECEPTOR WITH AND WITHOUT A LIGAND | | Descriptor: | 1,10-PHENANTHROLINE, ASPARTATE RECEPTOR | | Authors: | Kim, S.-H, Scott, W, Yeh, J.I, Prive, G.G, Milburn, M. | | Deposit date: | 1995-04-18 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand.
Science, 254, 1991
|
|
2LAO
 
 | | THREE-DIMENSIONAL STRUCTURES OF THE PERIPLASMIC LYSINE-, ARGININE-, ORNITHINE-BINDING PROTEIN WITH AND WITHOUT A LIGAND | | Descriptor: | LYSINE, ARGININE, ORNITHINE-BINDING PROTEIN | | Authors: | Kim, S.-H, Oh, B.-H, Kang, C.-H. | | Deposit date: | 1993-02-25 | | Release date: | 1994-06-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Three-dimensional structures of the periplasmic lysine/arginine/ornithine-binding protein with and without a ligand.
J.Biol.Chem., 268, 1993
|
|
1Q21
 
 | |
1LST
 
 | |
1LAG
 
 | | STRUCTURAL BASES FOR MULTIPLE LIGAND SPECIFICITY OF THE PERIPLASMIC LYSINE-, ARGININE-, ORNITHINE-BINDING PROTEIN | | Descriptor: | HISTIDINE, LYSINE, ARGININE, ... | | Authors: | Kim, S.-H, Oh, B.-H. | | Deposit date: | 1993-10-06 | | Release date: | 1995-07-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis for multiple ligand specificity of the periplasmic lysine-, arginine-, ornithine-binding protein.
J.Biol.Chem., 269, 1994
|
|
1LAF
 
 | | STRUCTURAL BASES FOR MULTIPLE LIGAND SPECIFICITY OF THE PERIPLASMIC LYSINE-, ARGININE-, ORNITHINE-BINDING PROTEIN | | Descriptor: | ARGININE, LYSINE, ORNITHINE-BINDING PROTEIN | | Authors: | Kim, S.-H, Oh, B.-H. | | Deposit date: | 1993-10-06 | | Release date: | 1995-07-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis for multiple ligand specificity of the periplasmic lysine-, arginine-, ornithine-binding protein.
J.Biol.Chem., 269, 1994
|
|
1WAT
 
 | |
1WAS
 
 | |
1THI
 
 | |
6TNA
 
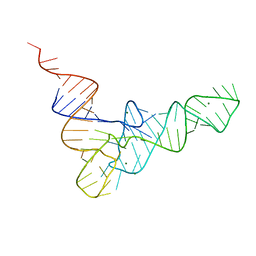 | | CRYSTAL STRUCTURE OF YEAST PHENYLALANINE T-RNA. I.CRYSTALLOGRAPHIC REFINEMENT | | Descriptor: | MAGNESIUM ION, TRNAPHE | | Authors: | Sussman, J.L, Holbrook, S.R, Warrant, R.W, Church, G.M, Kim, S.-H. | | Deposit date: | 1978-11-16 | | Release date: | 1979-01-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of yeast phenylalanine transfer RNA. I. Crystallographic refinement.
J.Mol.Biol., 123, 1978
|
|
2I1O
 
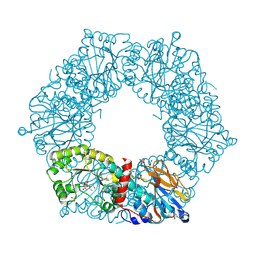 | | Crystal Structure of a Nicotinate Phosphoribosyltransferase from Thermoplasma acidophilum | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | Authors: | Shin, D.H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2006-08-14 | | Release date: | 2006-08-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of a zinc ion bound Nicotinate Phosphoribosyltransferase from Thermoplasma acidophilum
To be Published
|
|
2I14
 
 | |
1U3G
 
 | | Structural and Functional Characterization of a 5,10-Methenyltetrahydrofolate Synthetase from Mycoplasma pneumoniae (GI: 13508087) | | Descriptor: | 5,10-Methenyltetrahydrofolate Synthetase, 5-HYDROXYMETHYLENE-6-HYDROFOLIC ACID, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Chen, S, Yakunin, A.F, Proudfoot, M, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-07-21 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of methenyltetrahydrofolate synthetase from Mycoplasma pneumoniae (GI: 13508087) at 2.2 A resolution
Proteins, 56, 2004
|
|
1U8W
 
 | | Crystal structure of Arabidopsis thaliana nucleoside diphosphate kinase 1 | | Descriptor: | Nucleoside diphosphate kinase I | | Authors: | Im, Y.J, Kim, J.-I, Shen, Y, Na, Y, Han, Y.-J, Kim, S.-H, Song, P.-S, Eom, S.H. | | Deposit date: | 2004-08-07 | | Release date: | 2004-11-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural analysis of Arabidopsis thaliana nucleoside diphosphate kinase-2 for phytochrome-mediated light signaling
J.Mol.Biol., 343, 2004
|
|
