8WO8
 
 | | Crystal Structure of an RNA-binding protein, FAU-1, from Pyrococcus furiosus | | Descriptor: | Probable ribonuclease FAU-1, RNA (5'-R(P*AP*UP*A)-3') | | Authors: | Kawai, G, Okada, K, Baba, S, Sato, A, Sakamoto, T, Kanai, A. | | Deposit date: | 2023-10-06 | | Release date: | 2024-02-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Homo-trimeric structure of the ribonuclease for rRNA processing, FAU-1, from Pyrococcus furiosus.
J.Biochem., 175, 2024
|
|
2IP4
 
 | | Crystal Structure of Glycinamide Ribonucleotide Synthetase from Thermus thermophilus HB8 | | Descriptor: | Phosphoribosylamine--glycine ligase, SULFATE ION | | Authors: | Sampei, G, Baba, S, Kanagawa, M, Yanai, H, Ishii, T, Kawai, H, Fukai, Y, Ebihara, A, Nakagawa, N, Kawai, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-10-11 | | Release date: | 2007-10-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of glycinamide ribonucleotide synthetase, PurD, from thermophilic eubacteria
J.Biochem., 148, 2010
|
|
3W7B
 
 | | Crystal structure of formyltetrahydrofolate deformylase from Thermus thermophilus HB8 | | Descriptor: | Formyltetrahydrofolate deformylase | | Authors: | Sampei, G, Yanagida, Y, Ogata, N, Kusano, M, Terao, K, Kawai, H, Fukai, Y, Kanagawa, M, Inoue, Y, Baba, S, Kawai, G. | | Deposit date: | 2013-02-28 | | Release date: | 2014-01-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structures and reaction mechanisms of the two related enzymes, PurN and PurU
J.Biochem., 154, 2013
|
|
1WKS
 
 | |
8ZNQ
 
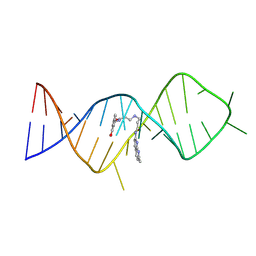 | | Solution structure of the complex of naphthyridine-azaquinolone and an RNA with ACG/AUA motif | | Descriptor: | N~3~-{3-[(7-METHYL-1,8-NAPHTHYRIDIN-2-YL)AMINO]-3-OXOPROPYL}-N~1~-[(7-OXO-7,8-DIHYDRO-1,8-NAPHTHYRIDIN-2-YL)METHYL]-BET A-ALANINAMIDE, RNA (30-MER) | | Authors: | Fujiwara, A, Chen, Q, Nakatani, K, Murata, A, Kawai, G. | | Deposit date: | 2024-05-27 | | Release date: | 2025-06-04 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the complex of naphthyridine-azaquinolone and an RNA with ACG/AUA motif
To Be Published
|
|
3HSB
 
 | | Crystal structure of YmaH (Hfq) from Bacillus subtilis in complex with an RNA aptamer | | Descriptor: | Protein hfq, RNA (5'-R(*AP*GP*AP*GP*AP*GP*A)-3') | | Authors: | Baba, S, Someya, T, Kumasaka, T, Kawai, G, Nakamura, K. | | Deposit date: | 2009-06-10 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Hfq from Bacillus subtilis in complex with SELEX-derived RNA aptamer: insight into RNA-binding properties of bacterial Hfq
Nucleic Acids Res., 40, 2012
|
|
6IZP
 
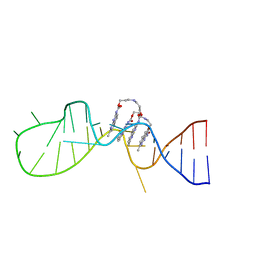 | | Solution structure of the complex of naphthyridine carbamate dimer and an RNA with UGGAA-UGGAA pentad | | Descriptor: | 3-[3-[(7-methyl-1,8-naphthyridin-2-yl)carbamoyloxy]propylamino]propyl ~{N}-(7-methyl-1,8-naphthyridin-2-yl)carbamate, RNA (29-MER) | | Authors: | Nagano, K, Shibata, T, Nakatani, K, Kawai, G. | | Deposit date: | 2018-12-20 | | Release date: | 2019-12-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Small molecule targeting r(UGGAA)n disrupts RNA foci and alleviates disease phenotype in Drosophila model
Nat Commun, 12, 2021
|
|
9U6Z
 
 | | INTERACTION BETWEEN A FLUOROQUINOLONE DERIVATIVE AND RNAS WITH A SINGLE BULGE | | Descriptor: | 1-cyclopropyl-N-[3-(dimethylamino)propyl]-7-(4-ethylpiperazin-1-yl)-6-fluoranyl-4-oxidanylidene-quinoline-3-carboxamide, DNA/RNA (5'-R(*GP*GP*AP*UP*GP*CP*UP*UP*UP*CP*GP*AP*GP*C)-D(P*C)-R(P*AP*UP*CP*C)-3') | | Authors: | Ichijo, R, Kawai, G. | | Deposit date: | 2025-03-23 | | Release date: | 2025-07-09 | | Method: | SOLUTION NMR | | Cite: | Interaction between a fluoroquinolone derivative KG022 and RNAs: effect of the bulged residues.
J.Biochem., 2025
|
|
9U70
 
 | | INTERACTION BETWEEN A FLUOROQUINOLONE DERIVATIVE AND RNAS WITH A SINGLE BULGE | | Descriptor: | 1-cyclopropyl-N-[3-(dimethylamino)propyl]-7-(4-ethylpiperazin-1-yl)-6-fluoranyl-4-oxidanylidene-quinoline-3-carboxamide, DNA/RNA (5'-R(*GP*GP*AP*UP*GP*CP*UP*UP*UP*CP*GP*AP*GP*C)-D(P*G)-R(P*AP*UP*CP*C)-3') | | Authors: | Ichijo, R, Kawai, G. | | Deposit date: | 2025-03-23 | | Release date: | 2025-07-09 | | Method: | SOLUTION NMR | | Cite: | Interaction between a fluoroquinolone derivative KG022 and RNAs: effect of the bulged residues.
J.Biochem., 2025
|
|
2FYH
 
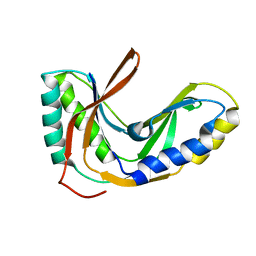 | | Solution structure of the 2'-5' RNA ligase-like protein from Pyrococcus furiosus | | Descriptor: | putative integral membrane transport protein | | Authors: | Okada, K, Matsuda, T, Sakamoto, T, Muto, Y, Yokoyama, S, Kanai, A, Kawai, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-02-08 | | Release date: | 2007-02-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Characterization of a heat-stable enzyme possessing GTP-dependent RNA ligase activity from a hyperthermophilic archaeon, Pyrococcus furiosus
Rna, 15, 2009
|
|
8I43
 
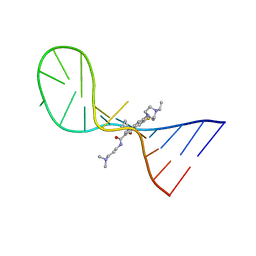 | |
8I44
 
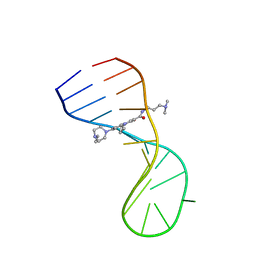 | |
8I45
 
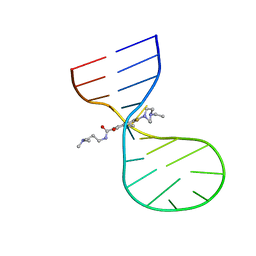 | |
8I46
 
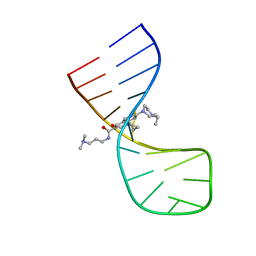 | |
8JHP
 
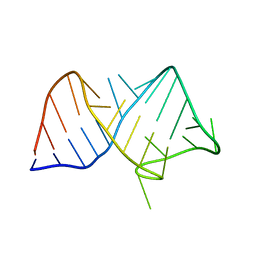 | |
8K8B
 
 | |
1L1W
 
 | | NMR structure of a SRP19 binding domain in human SRP RNA | | Descriptor: | SRP19 binding domain of SRP RNA | | Authors: | Sakamoto, T, Morita, S, Tabata, K, Nakamura, K, Kawai, G. | | Deposit date: | 2002-02-20 | | Release date: | 2002-05-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a SRP19 binding domain in human SRP RNA.
J.Biochem.(Tokyo), 132, 2002
|
|
9IO0
 
 | |
9IOU
 
 | |
9IOS
 
 | |
9IO1
 
 | |
9IOR
 
 | |
1K2G
 
 | | Structural basis for the 3'-terminal guanosine recognition by the group I intron | | Descriptor: | 5'-R(*CP*AP*GP*AP*CP*UP*UP*CP*GP*GP*UP*CP*GP*CP*AP*GP*AP*GP*AP*UP*GP*G)-3' | | Authors: | Kitamura, Y, Muto, Y, Watanabe, S, Kim, I, Ito, T, Nishiya, Y, Sakamoto, K, Ohtsuki, T, Kawai, G, Watanabe, K, Hosono, K, Takaku, H, Katoh, E, Yamazaki, T, Inoue, T, Yokoyama, S. | | Deposit date: | 2001-09-27 | | Release date: | 2002-05-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an RNA fragment with the P7/P9.0 region and the 3'-terminal guanosine of the tetrahymena group I intron.
RNA, 8, 2002
|
|
8JBD
 
 | |
2YRX
 
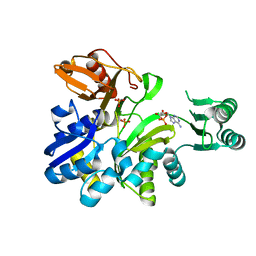 | | Crystal structure of GAR synthetase from Geobacillus kaustophilus | | Descriptor: | ADENOSINE MONOPHOSPHATE, PHOSPHATE ION, Phosphoribosylglycinamide synthetase | | Authors: | Baba, S, Kanagawa, M, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of glycinamide ribonucleotide synthetase, PurD, from thermophilic eubacteria
J.Biochem., 148, 2010
|
|
