1IST
 
 | |
7XGK
 
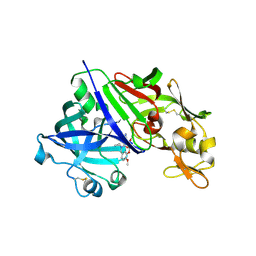 | | Human renin in complex with compound1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Renin, UNKNOWN LIGAND | | Authors: | Kashima, A. | | Deposit date: | 2022-04-05 | | Release date: | 2022-08-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of Novel 2-Carbamoyl Morpholine Derivatives as Highly Potent and Orally Active Direct Renin Inhibitors.
Acs Med.Chem.Lett., 13, 2022
|
|
7XGO
 
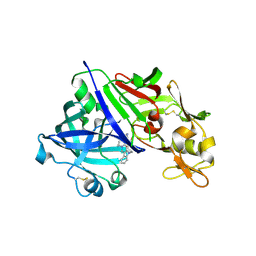 | | Human renin in complex with compound2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Renin, UNKNOWN LIGAND | | Authors: | Kashima, A. | | Deposit date: | 2022-04-05 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of Novel 2-Carbamoyl Morpholine Derivatives as Highly Potent and Orally Active Direct Renin Inhibitors.
Acs Med.Chem.Lett., 13, 2022
|
|
7XGP
 
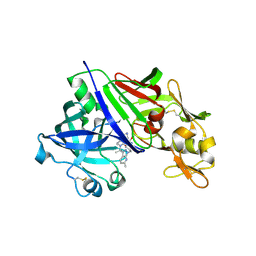 | | Human renin in complex with compound3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Renin, UNKNOWN LIGAND | | Authors: | Kashima, A. | | Deposit date: | 2022-04-05 | | Release date: | 2022-09-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery of SPH3127: A Novel, Highly Potent, and Orally Active Direct Renin Inhibitor.
J.Med.Chem., 65, 2022
|
|
1AB9
 
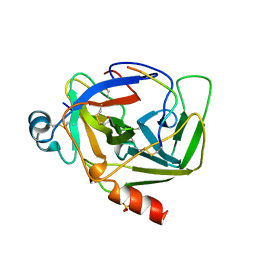 | | CRYSTAL STRUCTURE OF BOVINE GAMMA-CHYMOTRYPSIN | | Descriptor: | GAMMA-CHYMOTRYPSIN, PENTAPEPTIDE (TPGVY), SULFATE ION | | Authors: | Sugio, S, Kashima, A, Inoue, Y, Maeda, I, Nose, T, Shimohigashi, Y. | | Deposit date: | 1997-02-05 | | Release date: | 1997-08-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystal structure of a dipeptide-chymotrypsin complex in an inhibitory interaction.
Eur.J.Biochem., 255, 1998
|
|
1AFQ
 
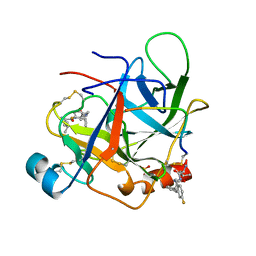 | | CRYSTAL STRUCTURE OF BOVINE GAMMA-CHYMOTRYPSIN COMPLEXED WITH A SYNTHETIC INHIBITOR | | Descriptor: | BOVINE GAMMA-CHYMOTRYPSIN, D-leucyl-N-(4-fluorobenzyl)-L-phenylalaninamide, SULFATE ION | | Authors: | Sugio, S, Kashima, A, Inoue, Y, Maeda, I, Nose, T, Shimohigashi, Y. | | Deposit date: | 1997-03-12 | | Release date: | 1997-09-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of a dipeptide-chymotrypsin complex in an inhibitory interaction.
Eur.J.Biochem., 255, 1998
|
|
8YVG
 
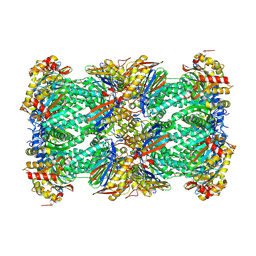 | | canine immunoproteasome 20S subunit in complex with compound 1 | | Descriptor: | Proteasome subunit alpha type, Proteasome subunit beta, Proteasome subunit beta type-8, ... | | Authors: | Kashima, A, Arai, Y. | | Deposit date: | 2024-03-28 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Optimization of alpha-amido boronic acids via cryo-electron microscopy analysis: Discovery of a novel highly selective immunoproteasome subunit LMP7 ( beta 5i)/LMP2 ( beta 1i) dual inhibitor.
Bioorg.Med.Chem., 109, 2024
|
|
8YSX
 
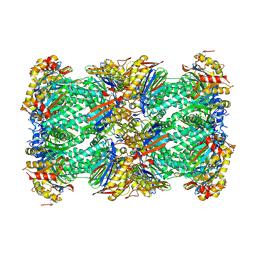 | | canine immunoproteasome 20S subunit in complex with compound 2 | | Descriptor: | Proteasome subunit alpha type, Proteasome subunit beta, Proteasome subunit beta type-8, ... | | Authors: | Kashima, A, Arai, Y. | | Deposit date: | 2024-03-24 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Optimization of alpha-amido boronic acids via cryo-electron microscopy analysis: Discovery of a novel highly selective immunoproteasome subunit LMP7 ( beta 5i)/LMP2 ( beta 1i) dual inhibitor.
Bioorg.Med.Chem., 109, 2024
|
|
8YVP
 
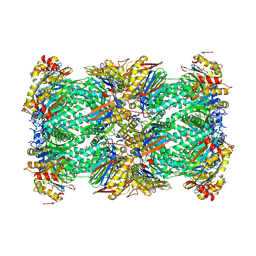 | | canine immunoproteasome 20S subunit in complex with compound 1 | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Kashima, A, Arai, Y. | | Deposit date: | 2024-03-29 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Optimization of alpha-amido boronic acids via cryo-electron microscopy analysis: Discovery of a novel highly selective immunoproteasome subunit LMP7 ( beta 5i)/LMP2 ( beta 1i) dual inhibitor.
Bioorg.Med.Chem., 109, 2024
|
|
8YPK
 
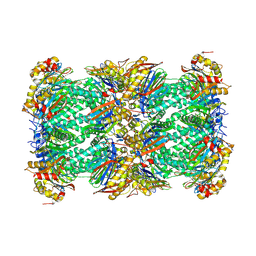 | | mouse proteasome 20S subunit in complex with compound 1 | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Kashima, A, Arai, Y. | | Deposit date: | 2024-03-17 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Optimization of alpha-amido boronic acids via cryo-electron microscopy analysis: Discovery of a novel highly selective immunoproteasome subunit LMP7 ( beta 5i)/LMP2 ( beta 1i) dual inhibitor.
Bioorg.Med.Chem., 109, 2024
|
|
1BM0
 
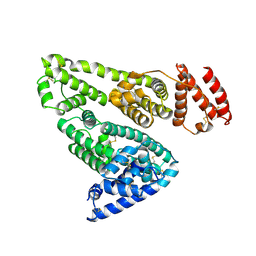 | | CRYSTAL STRUCTURE OF HUMAN SERUM ALBUMIN | | Descriptor: | SERUM ALBUMIN | | Authors: | Sugio, S, Kashima, A, Mochizuki, S, Noda, M, Kobayashi, K. | | Deposit date: | 1998-07-28 | | Release date: | 1999-07-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human serum albumin at 2.5 A resolution.
Protein Eng., 12, 1999
|
|
1WRM
 
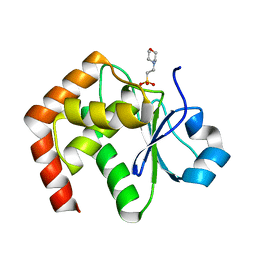 | | Crystal structure of JSP-1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, dual specificity phosphatase 22 | | Authors: | Yokota, T, Kashima, A, Kato, R, Sugio, S. | | Deposit date: | 2004-10-22 | | Release date: | 2005-10-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of human dual specificity phosphatase, JNK stimulatory phosphatase-1, at 1.5 A resolution
Proteins, 66, 2006
|
|
1A0G
 
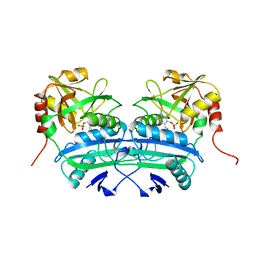 | | L201A MUTANT OF D-AMINO ACID AMINOTRANSFERASE COMPLEXED WITH PYRIDOXAMINE-5'-PHOSPHATE | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, D-AMINO ACID AMINOTRANSFERASE | | Authors: | Sugio, S, Kashima, A, Kishimoto, K, Peisach, D, Petsko, G.A, Ringe, D, Yoshimura, T, Esaki, N. | | Deposit date: | 1997-11-30 | | Release date: | 1998-06-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of L201A mutant of D-amino acid aminotransferase at 2.0 A resolution: implication of the structural role of Leu201 in transamination.
Protein Eng., 11, 1998
|
|
2DAB
 
 | | L201A MUTANT OF D-AMINO ACID AMINOTRANSFERASE COMPLEXED WITH PYRIDOXAL-5'-PHOSPHATE | | Descriptor: | D-AMINO ACID AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Sugio, S, Kashima, A, Kishimoto, K, Peisach, D, Petsko, G.A, Ringe, D, Yoshimura, T, Esaki, N. | | Deposit date: | 1997-11-30 | | Release date: | 1998-06-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of L201A mutant of D-amino acid aminotransferase at 2.0 A resolution: implication of the structural role of Leu201 in transamination.
Protein Eng., 11, 1998
|
|
1AO6
 
 | |
2E1M
 
 | | Crystal Structure of L-Glutamate Oxidase from Streptomyces sp. X-119-6 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-glutamate oxidase, PHOSPHATE ION | | Authors: | Sasaki, C, Kashima, A, Sakaguchi, C, Mizuno, H, Arima, J, Kusakabe, H, Tamura, T, Sugio, S, Inagaki, K. | | Deposit date: | 2006-10-26 | | Release date: | 2007-11-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural characterization of l-glutamate oxidase from Streptomyces sp. X-119-6
Febs J., 276, 2009
|
|
1QCM
 
 | | AMYLOID BETA PEPTIDE (25-35), NMR, 20 STRUCTURES | | Descriptor: | AMYLOID BETA PEPTIDE | | Authors: | Kohno, T, Kobayashi, K, Maeda, T, Sato, K, Takashima, A. | | Deposit date: | 1996-07-19 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structures of the amyloid beta peptide (25-35) in membrane-mimicking environment.
Biochemistry, 35, 1996
|
|
4YVF
 
 | | Structure of S-adenosyl-L-homocysteine hydrolase | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-{[5-chloro-2-(4-chlorophenoxy)phenyl](2-{[2-(methylamino)ethyl]amino}-2-oxoethyl)amino}-N-(1,3-dihydro-2H-isoindol-2-yl)-N-methylacetamide, Adenosylhomocysteinase | | Authors: | Akiko, K. | | Deposit date: | 2015-03-20 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery and structural analyses of S-adenosyl-L-homocysteine hydrolase inhibitors based on non-adenosine analogs.
Bioorg.Med.Chem., 23, 2015
|
|
5LAE
 
 | | Crystal structure of murine N1-acetylpolyamine oxidase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Peroxisomal N(1)-acetyl-spermine/spermidine oxidase,Peroxisomal N(1)-acetyl-spermine/spermidine oxidase | | Authors: | Sjogren, T, Aagaard, A, Snijder, A, Barlind, L. | | Deposit date: | 2016-06-14 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Structure of Murine N(1)-Acetylspermine Oxidase Reveals Molecular Details of Vertebrate Polyamine Catabolism.
Biochemistry, 56, 2017
|
|
5MBX
 
 | | Crystal structure of reduced murine N1-acetylpolyamine oxidase in complex with N1-acetylspermine | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, N-[3-({4-[(3-aminopropyl)amino]butyl}amino)propyl]acetamide, Peroxisomal N(1)-acetyl-spermine/spermidine oxidase, ... | | Authors: | Sjogren, T, Aagaard, A, Wassvik, C, Snijder, A, Barlind, L. | | Deposit date: | 2016-11-09 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Structure of Murine N(1)-Acetylspermine Oxidase Reveals Molecular Details of Vertebrate Polyamine Catabolism.
Biochemistry, 56, 2017
|
|
5LFO
 
 | | Crystal structure of murine N1-acetylpolyamine oxidase in complex with N1-acetylspermine | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, N-[3-({4-[(3-aminopropyl)amino]butyl}amino)propyl]acetamide, ... | | Authors: | Sjogren, T, Aagaard, A, Wassvik, C, Snijder, A, Barlind, L. | | Deposit date: | 2016-07-04 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | The Structure of Murine N(1)-Acetylspermine Oxidase Reveals Molecular Details of Vertebrate Polyamine Catabolism.
Biochemistry, 56, 2017
|
|
5LGB
 
 | | Crystal structure of murine N1-acetylpolyamine oxidase in complex with MDL72527 | | Descriptor: | FAD-MDL72527 adduct, N,N'-BIS(2,3-BUTADIENYL)-1,4-BUTANE-DIAMINE, Peroxisomal N(1)-acetyl-spermine/spermidine oxidase,Peroxisomal N(1)-acetyl-spermine/spermidine oxidase | | Authors: | Sjogren, T, Aagaard, A, Wassvik, C, Snijder, A, Barlind, L. | | Deposit date: | 2016-07-06 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of Murine N(1)-Acetylspermine Oxidase Reveals Molecular Details of Vertebrate Polyamine Catabolism.
Biochemistry, 56, 2017
|
|
5CH4
 
 | | Peptide-Bound State of Thermus thermophilus SecYEG | | Descriptor: | Protein translocase subunit SecE, Protein translocase subunit SecY, Putative preprotein translocase, ... | | Authors: | Tanaka, Y, Sugano, Y, Takemoto, M, Kusakizako, T, Kumazaki, K, Ishitani, R, Nureki, O, Tsukazaki, T. | | Deposit date: | 2015-07-10 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.64 Å) | | Cite: | Crystal Structures of SecYEG in Lipidic Cubic Phase Elucidate a Precise Resting and a Peptide-Bound State.
Cell Rep, 13, 2015
|
|
5AWW
 
 | | Precise Resting State of Thermus thermophilus SecYEG | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Protein translocase subunit SecE, Protein translocase subunit SecY, ... | | Authors: | Tanaka, Y, Sugano, Y, Takemoto, M, Kusakizako, T, Kumazaki, K, Ishitani, R, Nureki, O, Tsukazaki, T. | | Deposit date: | 2015-07-10 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.724 Å) | | Cite: | Crystal Structures of SecYEG in Lipidic Cubic Phase Elucidate a Precise Resting and a Peptide-Bound State.
Cell Rep, 13, 2015
|
|
8XUK
 
 | | Structure of beta-1,2-glucanase from Photobacterium gaetbulicola (PgSGL3, ligand-free) | | Descriptor: | CHLORIDE ION, H744_1c0222 | | Authors: | Nakajima, M, Motouchi, S, Nakai, H. | | Deposit date: | 2024-01-13 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | beta-1,2-Glucanase superfamily identified by sequential, functional and structural analyses
To Be Published
|
|
