7NVG
 
 | | Salmonella flagellar basal body refined in C1 map | | Descriptor: | Basal-body rod modification protein FlgD, Flagellar L-ring protein, Flagellar M-ring protein, ... | | Authors: | Johnson, S, Furlong, E, Lea, S.M. | | Deposit date: | 2021-03-15 | | Release date: | 2021-05-05 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular structure of the intact bacterial flagellar basal body.
Nat Microbiol, 6, 2021
|
|
6R69
 
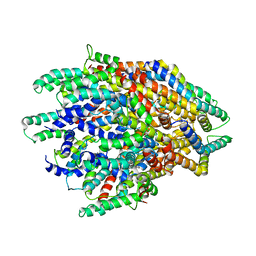 | | Improved map of the FliPQR complex that forms the core of the Salmonella type III secretion system export apparatus. | | Descriptor: | Flagellar biosynthetic protein FliP, Flagellar biosynthetic protein FliQ, Flagellar biosynthetic protein FliR | | Authors: | Johnson, S, Kuhlen, L, Abrusci, P, Lea, S.M. | | Deposit date: | 2019-03-26 | | Release date: | 2019-05-29 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | The Structure of an Injectisome Export Gate Demonstrates Conservation of Architecture in the Core Export Gate between Flagellar and Virulence Type III Secretion Systems.
Mbio, 10, 2019
|
|
6R6B
 
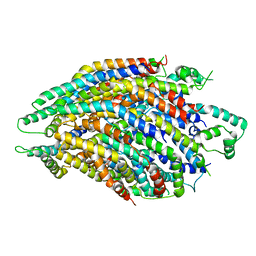 | | Structure of the core Shigella flexneri type III secretion system export gate complex SctRST (Spa24/Spa9/Spa29). | | Descriptor: | Surface presentation of antigens protein SpaP, Surface presentation of antigens protein SpaQ, Surface presentation of antigens protein SpaR | | Authors: | Johnson, S, Kuhlen, L, Deme, J.C, Abrusci, P, Lea, S.M. | | Deposit date: | 2019-03-26 | | Release date: | 2019-05-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The Structure of an Injectisome Export Gate Demonstrates Conservation of Architecture in the Core Export Gate between Flagellar and Virulence Type III Secretion Systems.
Mbio, 10, 2019
|
|
7BK0
 
 | |
7BJ2
 
 | |
7BGL
 
 | | Salmonella LP ring 26 mer refined in C26 map | | Descriptor: | (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-4,5-bis(oxidanyl)oxane-2-carboxylic acid, Flagellar L-ring protein, Flagellar P-ring protein, ... | | Authors: | Johnson, S, Furlong, E, Lea, S.M. | | Deposit date: | 2021-01-07 | | Release date: | 2021-05-05 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Molecular structure of the intact bacterial flagellar basal body.
Nat Microbiol, 6, 2021
|
|
7BHQ
 
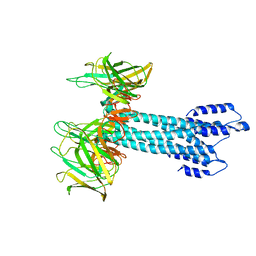 | |
7BIN
 
 | | Salmonella export gate and rod refined in focussed C1 map | | Descriptor: | Flagellar basal body rod protein FlgB, Flagellar basal-body rod protein FlgC, Flagellar basal-body rod protein FlgF, ... | | Authors: | Johnson, S, Furlong, E, Lea, S.M. | | Deposit date: | 2021-01-12 | | Release date: | 2021-05-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular structure of the intact bacterial flagellar basal body.
Nat Microbiol, 6, 2021
|
|
4AYM
 
 | | Structure of a complex between CCPs 6 and 7 of Human Complement Factor H and Neisseria meningitidis FHbp Variant 3 P106A mutant | | Descriptor: | COMPLEMENT FACTOR H, FACTOR H BINDING PROTEIN | | Authors: | Johnson, S, Tan, L, van der Veen, S, Caesar, J, Goicoechea De Jorge, E, Everett, R.J, Bai, X, Exley, R.M, Ward, P.N, Ruivo, N, Trivedi, K, Cumber, E, Jones, R, Newham, L, Staunton, D, Borrow, R, Pickering, M, Lea, S.M, Tang, C.M. | | Deposit date: | 2012-06-21 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Design and Evaluation of Meningococcal Vaccines Through Structure-Based Modification of Host and Pathogen Molecules.
Plos Pathog., 8, 2012
|
|
4AYD
 
 | | Structure of a complex between CCPs 6 and 7 of Human Complement Factor H and Neisseria meningitidis FHbp Variant 1 R106A mutant | | Descriptor: | 1,2-ETHANEDIOL, COMPLEMENT FACTOR H, FACTOR H BINDING PROTEIN | | Authors: | Johnson, S, Tan, L, van der Veen, S, Caesar, J, Goicoechea De Jorge, E, Everett, R.J, Bai, X, Exley, R.M, Ward, P.N, Ruivo, N, Trivedi, K, Cumber, E, Jones, R, Newham, L, Staunton, D, Borrow, R, Pickering, M, Lea, S.M, Tang, C.M. | | Deposit date: | 2012-06-20 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and Evaluation of Meningococcal Vaccines Through Structure-Based Modification of Host and Pathogen Molecules
Plos Pathog., 8, 2012
|
|
4AYI
 
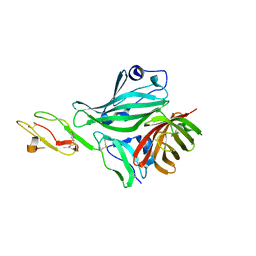 | | Structure of a complex between CCPs 6 and 7 of Human Complement Factor H and Neisseria meningitidis FHbp Variant 3 Wild type | | Descriptor: | 1,2-ETHANEDIOL, COMPLEMENT FACTOR H, LIPOPROTEIN GNA1870 CCOMPND 7 | | Authors: | Johnson, S, Tan, L, van der Veen, S, Caesar, J, Goicoechea De Jorge, E, Everett, R.J, Bai, X, Exley, R.M, Ward, P.N, Ruivo, N, Trivedi, K, Cumber, E, Jones, R, Newham, L, Staunton, D, Borrow, R, Pickering, M, Lea, S.M, Tang, C.M. | | Deposit date: | 2012-06-21 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Design and Evaluation of Meningococcal Vaccines Through Structure-Based Modification of Host and Pathogen Molecules.
Plos Pathog., 8, 2012
|
|
4AYN
 
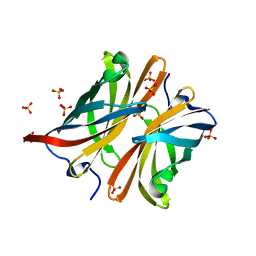 | | Structure of the C-terminal barrel of Neisseria meningitidis FHbp Variant 2 | | Descriptor: | FACTOR H-BINDING PROTEIN, SULFATE ION | | Authors: | Johnson, S, Tan, L, van der Veen, S, Caesar, J, Goicoechea De Jorge, E, Everett, R.J, Bai, X, Exley, R.M, Ward, P.N, Ruivo, N, Trivedi, K, Cumber, E, Jones, R, Newham, L, Staunton, D, Borrow, R, Pickering, M, Lea, S.M, Tang, C.M. | | Deposit date: | 2012-06-21 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Design and Evaluation of Meningococcal Vaccines Through Structure-Based Modification of Host and Pathogen Molecules.
Plos Pathog., 8, 2012
|
|
4AYE
 
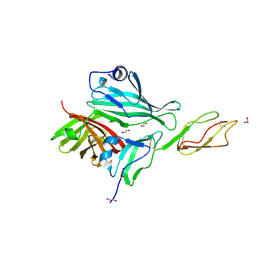 | | Structure of a complex between CCPs 6 and 7 of Human Complement Factor H and Neisseria meningitidis FHbp Variant 1 E283AE304A mutant | | Descriptor: | 1,2-ETHANEDIOL, COMPLEMENT FACTOR H, FACTOR H BINDING PROTEIN | | Authors: | Johnson, S, Tan, L, van der Veen, S, Caesar, J, Goicoechea De Jorge, E, Everett, R.J, Bai, X, Exley, R.M, Ward, P.N, Ruivo, N, Trivedi, K, Cumber, E, Jones, R, Newham, L, Staunton, D, Borrow, R, Pickering, M, Lea, S.M, Tang, C.M. | | Deposit date: | 2012-06-20 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Design and Evaluation of Meningococcal Vaccines Through Structure-Based Modification of Host and Pathogen Molecules.
Plos Pathog., 8, 2012
|
|
8UPL
 
 | | Cryo-EM structure of a Clockwise locked form of the Salmonella enterica Typhimurium flagellar C-ring, with C34 symmetry applied | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG, Flagellar motor switch protein FliM, ... | | Authors: | Johnson, S, Deme, J.C, Lea, S.M. | | Deposit date: | 2023-10-22 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Structural basis of directional switching by the bacterial flagellum.
Nat Microbiol, 9, 2024
|
|
8UOX
 
 | | Cryo-EM structure of a Counterclockwise locked form of the Salmonella enterica Typhimurium flagellar C-ring, with C34 symmetry applied | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG, Flagellar motor switch protein FliM, ... | | Authors: | Johnson, S, Deme, J.C, Lea, S.M. | | Deposit date: | 2023-10-20 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis of directional switching by the bacterial flagellum.
Nat Microbiol, 9, 2024
|
|
8UMD
 
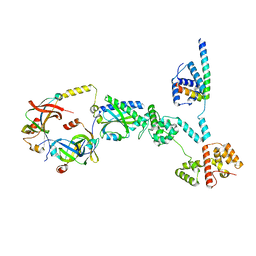 | | Cryo-EM structure of a single subunit of a Counterclockwise-locked form of the Salmonella enterica Typhimurium flagellar C-ring. | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG, Flagellar motor switch protein FliM, ... | | Authors: | Johnson, S, Deme, J.C, Lea, S.M. | | Deposit date: | 2023-10-17 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of directional switching by the bacterial flagellum.
Nat Microbiol, 9, 2024
|
|
8UMX
 
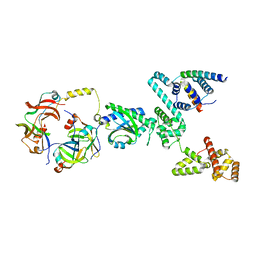 | | Cryo-EM structure of a single subunit of a Clockwise-locked form of the Salmonella enterica Typhimurium flagellar C-ring. | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG, Flagellar motor switch protein FliM, ... | | Authors: | Johnson, S, Deme, J.C, Lea, S.M. | | Deposit date: | 2023-10-18 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of directional switching by the bacterial flagellum.
Nat Microbiol, 9, 2024
|
|
6SD1
 
 | | Structure of the RBM3/collar region of the Salmonella flagella MS-ring protein FliF with 33-fold symmetry applied | | Descriptor: | Flagellar M-ring protein | | Authors: | Johnson, S, Fong, Y.H, Deme, J.C, Furlong, E.J, Kuhlen, L, Lea, S.M. | | Deposit date: | 2019-07-26 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Symmetry mismatch in the MS-ring of the bacterial flagellar rotor explains the structural coordination of secretion and rotation.
Nat Microbiol, 5, 2020
|
|
6SCN
 
 | | 33mer structure of the Salmonella flagella MS-ring protein FliF | | Descriptor: | Flagellar M-ring protein | | Authors: | Johnson, S, Fong, Y.H, Deme, J.C, Furlong, E.J, Kuhlen, L, Lea, S.M. | | Deposit date: | 2019-07-24 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Symmetry mismatch in the MS-ring of the bacterial flagellar rotor explains the structural coordination of secretion and rotation.
Nat Microbiol, 5, 2020
|
|
6SD3
 
 | | 34mer structure of the Salmonella flagella MS-ring protein FliF | | Descriptor: | Flagellar M-ring protein | | Authors: | Johnson, S, Fong, Y.H, Deme, J.C, Furlong, E.J, Kuhlen, L, Lea, S.M. | | Deposit date: | 2019-07-26 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Symmetry mismatch in the MS-ring of the bacterial flagellar rotor explains the structural coordination of secretion and rotation.
Nat Microbiol, 5, 2020
|
|
6SD2
 
 | | Structure of the RBM2inner region of the Salmonella flagella MS-ring protein FliF with 21-fold symmetry applied. | | Descriptor: | Flagellar M-ring protein | | Authors: | Johnson, S, Fong, Y.H, Deme, J.C, Furlong, E.J, Kuhlen, L, Lea, S.M. | | Deposit date: | 2019-07-26 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Symmetry mismatch in the MS-ring of the bacterial flagellar rotor explains the structural coordination of secretion and rotation.
Nat Microbiol, 5, 2020
|
|
4BIK
 
 | | Structure of a disulfide locked mutant of Intermedilysin with human CD59 | | Descriptor: | CD59 GLYCOPROTEIN, INTERMEDILYSIN | | Authors: | Johnson, S, Brooks, N.J, Smith, R.A.G, Lea, S.M, Bubeck, D. | | Deposit date: | 2013-04-10 | | Release date: | 2013-05-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.494 Å) | | Cite: | Structural Basis for Recognition of the Pore- Forming Toxin Intermedilysin by Human Complement Receptor Cd59
Cell Rep., 3, 2013
|
|
6SD4
 
 | | Structure of the RBM3/collar region of the Salmonella flagella MS-ring protein FliF with 34-fold symmetry applied | | Descriptor: | Flagellar M-ring protein | | Authors: | Johnson, S, Fong, Y.H, Deme, J.C, Furlong, E.J, Kuhlen, L, Lea, S.M. | | Deposit date: | 2019-07-26 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Symmetry mismatch in the MS-ring of the bacterial flagellar rotor explains the structural coordination of secretion and rotation.
Nat Microbiol, 5, 2020
|
|
6SD5
 
 | | Structure of the RBM2 inner ring of Salmonella flagella MS-ring protein FliF with 22-fold symmetry applied | | Descriptor: | Flagellar M-ring protein | | Authors: | Johnson, S, Fong, Y.H, Deme, J.C, Furlong, E.J, Kuhlen, L, Lea, S.M. | | Deposit date: | 2019-07-26 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Symmetry mismatch in the MS-ring of the bacterial flagellar rotor explains the structural coordination of secretion and rotation.
Nat Microbiol, 5, 2020
|
|
6TRE
 
 | | Structure of the RBM3/collar region of the Salmonella flagella MS-ring protein FliF with 32-fold symmetry applied | | Descriptor: | Flagellar M-ring protein | | Authors: | Johnson, S, Fong, Y.H, Deme, J.C, Furlong, E.J, Kuhlen, L, Lea, S.M. | | Deposit date: | 2019-12-18 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the bacterial flagellar rotor MS-ring: a minimum inventory/maximum diversity system.
To Be Published
|
|
