6Z3N
 
 | |
6Z3O
 
 | |
1IHI
 
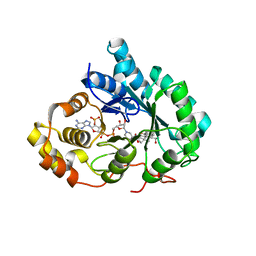 | | Crystal Structure of Human Type III 3-alpha-Hydroxysteroid Dehydrogenase/Bile Acid Binding Protein (AKR1C2) Complexed with NADP+ and Ursodeoxycholate | | Descriptor: | 3-ALPHA-HYDROXYSTEROID DEHYDROGENASE, ISO-URSODEOXYCHOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jin, Y, Stayrook, S.E, Albert, R.H, Palackal, N.T, Penning, T.M, Lewis, M. | | Deposit date: | 2001-04-19 | | Release date: | 2001-10-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human type III 3alpha-hydroxysteroid dehydrogenase/bile acid binding protein complexed with NADP(+) and ursodeoxycholate.
Biochemistry, 40, 2001
|
|
7P1Y
 
 | |
5DDT
 
 | |
5DDV
 
 | |
5JTS
 
 | | Structure of a beta-1,4-mannanase, SsGH134. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Jin, Y, Petricevic, M, Goddard-Borger, E.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2016-05-09 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | A beta-Mannanase with a Lysozyme-like Fold and a Novel Molecular Catalytic Mechanism.
ACS Cent Sci, 2, 2016
|
|
5JUG
 
 | | Structure of an inactive (E45Q) variant of a beta-1,4-mannanase, SsGH134, in complex with Man5 | | Descriptor: | CHLORIDE ION, GLYCEROL, alpha-D-mannopyranose, ... | | Authors: | Jin, Y, Petricevic, M, Goddard-Borger, E.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2016-05-10 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | A beta-Mannanase with a Lysozyme-like Fold and a Novel Molecular Catalytic Mechanism.
ACS Cent Sci, 2, 2016
|
|
5JU9
 
 | | Structure of a beta-1,4-mannanase, SsGH134, in complex with Man3. | | Descriptor: | CHLORIDE ION, beta-1,4-mannanase, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose | | Authors: | Jin, Y, Petricevic, M, Goddard-Borger, E.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2016-05-10 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | A beta-Mannanase with a Lysozyme-like Fold and a Novel Molecular Catalytic Mechanism.
ACS Cent Sci, 2, 2016
|
|
5G0M
 
 | | beta-glucuronidase with an activity-based probe (N-acyl cyclophellitol aziridine) bound | | Descriptor: | (1S,2R,3R,4S,6S)-6-[(8-azidooctanoyl)amino]-2,3,4-trihydroxycyclohexane-1-carboxylate, BETA-GLUCURONIDASE, PHOSPHATE ION | | Authors: | Jin, Y, Wu, L, Jiang, J.B, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2016-03-21 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of a Bacteria Beta-Glucuronidase in Gh79 Family
To be Published
|
|
5HS2
 
 | |
8JJA
 
 | | SP1746 in complex with acetate ions | | Descriptor: | ACETATE ION, FE (III) ION, bis(5'-nucleosyl)-tetraphosphatase (symmetrical) | | Authors: | Jin, Y, Niu, L, Ke, J. | | Deposit date: | 2023-05-30 | | Release date: | 2024-05-08 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and biochemical characterization of a nucleotide hydrolase from Streptococcus pneumonia.
Structure, 32, 2024
|
|
8JK8
 
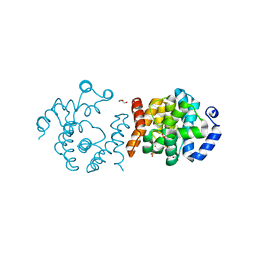 | | SP1746 in complex with UDP | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, GLYCEROL, ... | | Authors: | Jin, Y, Niu, L, Ke, J. | | Deposit date: | 2023-06-01 | | Release date: | 2024-05-08 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and biochemical characterization of a nucleotide hydrolase from Streptococcus pneumonia.
Structure, 32, 2024
|
|
8JKP
 
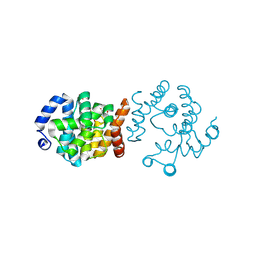 | | SP1746 in complex with dTMP | | Descriptor: | FE (III) ION, GLYCEROL, SULFATE ION, ... | | Authors: | Jin, Y, Ke, J, Niu, L. | | Deposit date: | 2023-06-01 | | Release date: | 2024-05-08 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and biochemical characterization of a nucleotide hydrolase from Streptococcus pneumonia.
Structure, 32, 2024
|
|
8JKR
 
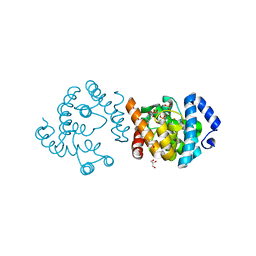 | | SP1746 in complex with UMP | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, GLYCEROL, ... | | Authors: | Jin, Y, Niu, L, Ke, J. | | Deposit date: | 2023-06-01 | | Release date: | 2024-05-08 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and biochemical characterization of a nucleotide hydrolase from Streptococcus pneumonia.
Structure, 32, 2024
|
|
8JKA
 
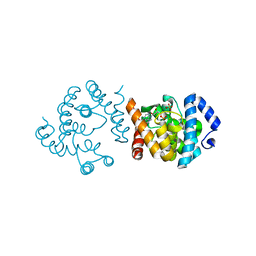 | | SP1746 in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, FE (III) ION, SULFATE ION, ... | | Authors: | Jin, Y, Niu, L, Ke, J. | | Deposit date: | 2023-06-01 | | Release date: | 2024-05-08 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and biochemical characterization of a nucleotide hydrolase from Streptococcus pneumonia.
Structure, 32, 2024
|
|
8JK5
 
 | | Crystal structure of SP1746 from Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) | | Descriptor: | FE (III) ION, GLYCEROL, SULFATE ION, ... | | Authors: | Jin, Y, Niu, L, Ke, J. | | Deposit date: | 2023-05-31 | | Release date: | 2024-05-08 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biochemical characterization of a nucleotide hydrolase from Streptococcus pneumonia.
Structure, 32, 2024
|
|
8JJK
 
 | | SP1746 in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FE (III) ION, GLYCEROL, ... | | Authors: | Jin, Y, Niu, L, Ke, J. | | Deposit date: | 2023-05-30 | | Release date: | 2024-05-08 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biochemical characterization of a nucleotide hydrolase from Streptococcus pneumonia.
Structure, 32, 2024
|
|
8JK9
 
 | | SP1746 in complex with GDP | | Descriptor: | FE (III) ION, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Jin, Y, Ke, J, Niu, L. | | Deposit date: | 2023-06-01 | | Release date: | 2024-05-08 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and biochemical characterization of a nucleotide hydrolase from Streptococcus pneumonia.
Structure, 32, 2024
|
|
8YT5
 
 | | SP1746 treated with EDTA, in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FE (III) ION, bis(5'-nucleosyl)-tetraphosphatase (symmetrical) | | Authors: | Jin, Y, Niu, L, Ke, J. | | Deposit date: | 2024-03-25 | | Release date: | 2024-05-08 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and biochemical characterization of a nucleotide hydrolase from Streptococcus pneumonia.
Structure, 32, 2024
|
|
7Z0L
 
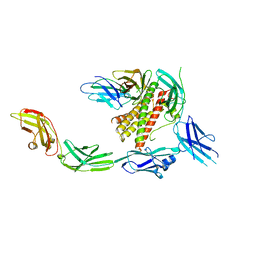 | | IL-27 signalling complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-27 receptor subunit alpha, ... | | Authors: | Jin, Y, Gardner, S, Bubeck, D. | | Deposit date: | 2022-02-23 | | Release date: | 2022-07-27 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into the assembly and activation of the IL-27 signaling complex.
Embo Rep., 23, 2022
|
|
5FOO
 
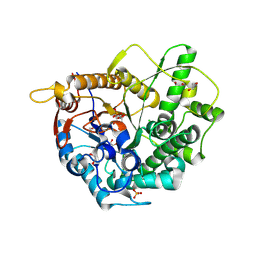 | | 6-phospho-beta-glucosidase | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-PHOSPHO-BETA-D GLYCOSIDASE, ... | | Authors: | Jin, Y, Kwan, D.H, Withers, S.G, Davies, G.J. | | Deposit date: | 2015-11-24 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Chemoenzymatic Synthesis of 6-Phospho-Cyclophellitol as a Novel Probe of 6-Phospho-Beta-Glucosidases.
FEBS Lett., 590, 2016
|
|
5G0Q
 
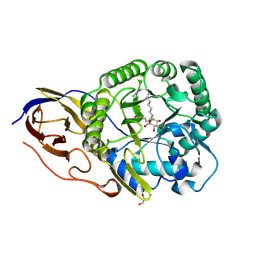 | | beta-glucuronidase with an activity-based probe (N-alkyl cyclophellitol aziridine) bound | | Descriptor: | (1R,2S,3R,4S,5S,6R)-7-(8-AZIDOOCTYL)-3,4,5-TRIHYDROXY-, BETA-GLUCURONIDASE, GLYCEROL | | Authors: | Jin, Y, Wu, L, Jiang, J.B, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2016-03-22 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Activity-based probes for functional interrogation of retaining beta-glucuronidases.
Nat. Chem. Biol., 13, 2017
|
|
5OHT
 
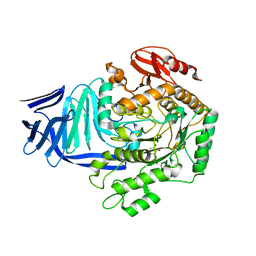 | | A GH31 family sulfoquinovosidase from E. coli in complex with aza-sugar inhibitor IFGSQ | | Descriptor: | CALCIUM ION, Sulfoquinovosidase, [(3~{S},4~{R},5~{R})-4,5-bis(oxidanyl)piperidin-3-yl]methanesulfonic acid | | Authors: | Jin, Y, Williams, S.J, Goddard-Borger, E, Davies, G.J. | | Deposit date: | 2017-07-18 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural and Biochemical Insights into the Function and Evolution of Sulfoquinovosidases.
ACS Cent Sci, 4, 2018
|
|
5OHS
 
 | | A GH31 family sulfoquinovosidase mutant D455N in complex with pNPSQ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 4-nitrophenyl alpha-D-6-sulfoquinovoside, ... | | Authors: | Jin, Y, Williams, S.J, Goddard-Borger, E, Davies, G.J. | | Deposit date: | 2017-07-18 | | Release date: | 2018-08-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and Biochemical Insights into the Function and Evolution of Sulfoquinovosidases.
ACS Cent Sci, 4, 2018
|
|
