1AZX
 
 | | ANTITHROMBIN/PENTASACCHARIDE COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,4-di-O-methyl-2,6-di-O-sulfo-alpha-D-glucopyranose-(1-4)-2,3-di-O-methyl-beta-D-glucopyranuronic acid-(1-4)-2,3,6-tri-O-sulfo-alpha-D-glucopyranose-(1-4)-3-O-methyl-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-methyl 2,3,6-tri-O-sulfo-alpha-D-glucopyranoside, ANTITHROMBIN | | Authors: | Jin, L, Abrahams, J.P, Skinner, R, Petitou, M, Pike, R.N, Carrell, R.W. | | Deposit date: | 1997-11-23 | | Release date: | 1999-01-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The anticoagulant activation of antithrombin by heparin.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1F1B
 
 | | CRYSTAL STRUCTURE OF E. COLI ASPARTATE TRANSCARBAMOYLASE P268A MUTANT IN THE R-STATE IN THE PRESENCE OF N-PHOSPHONACETYL-L-ASPARTATE | | Descriptor: | ASPARTATE CARBAMOYLTRANSFERASE CATALYTIC CHAIN, ASPARTATE CARBAMOYLTRANSFERASE REGULATORY CHAIN, N-(PHOSPHONACETYL)-L-ASPARTIC ACID, ... | | Authors: | Jin, L, Stec, B, Kantrowitz, E.R. | | Deposit date: | 2000-05-18 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A cis-proline to alanine mutant of E. coli aspartate transcarbamoylase: kinetic studies and three-dimensional crystal structures.
Biochemistry, 39, 2000
|
|
1U59
 
 | | Crystal Structure of the ZAP-70 Kinase Domain in Complex with Staurosporine | | Descriptor: | STAUROSPORINE, Tyrosine-protein kinase ZAP-70 | | Authors: | Jin, L, Pluskey, S, Petrella, E.C, Cantin, S.M, Gorga, J.C, Rynkiewicz, M.J, Pandey, P, Strickler, J.E, Babine, R.E, Weaver, D.T, Seidl, K.J. | | Deposit date: | 2004-07-27 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Three-dimensional Structure of the ZAP-70 Kinase Domain in Complex with Staurosporine: IMPLICATIONS FOR THE DESIGN OF SELECTIVE INHIBITORS
J.Biol.Chem., 279, 2004
|
|
1D09
 
 | | ASPARTATE TRANSCARBAMOYLASE COMPLEXED WITH N-PHOSPHONACETYL-L-ASPARTATE (PALA) | | Descriptor: | ASPARTATE CARBAMOYLTRANSFERASE CATALYTIC CHAIN, ASPARTATE CARBAMOYLTRANSFERASE REGULATORY CHAIN, N-(PHOSPHONACETYL)-L-ASPARTIC ACID, ... | | Authors: | Jin, L, Stec, B, Lipscomb, W.N, Kantrowitz, E.R. | | Deposit date: | 1999-09-09 | | Release date: | 2000-01-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights into the mechanisms of catalysis and heterotropic regulation of Escherichia coli aspartate transcarbamoylase based upon a structure of the enzyme complexed with the bisubstrate analogue N-phosphonacetyl-L-aspartate at 2.1 A.
Proteins, 37, 1999
|
|
1PZU
 
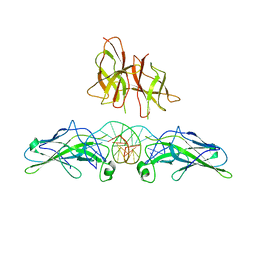 | | An asymmetric NFAT1-RHR homodimer on a pseudo-palindromic, Kappa-B site | | Descriptor: | 5'-D(*AP*AP*TP*GP*GP*AP*AP*AP*TP*TP*CP*CP*TP*C)-3', 5'-D(*TP*TP*GP*AP*GP*GP*AP*AP*TP*TP*TP*CP*CP*A)-3', Nuclear factor of activated T-cells, ... | | Authors: | Jin, L, Sliz, P, Chen, L, Macian, F, Rao, A, Hogan, P.G, Harrison, S.C. | | Deposit date: | 2003-07-14 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | An asymmetric NFAT1 dimer on a pseudo-palindromic KB-like DNA site
Nat.Struct.Biol., 10, 2003
|
|
1ET1
 
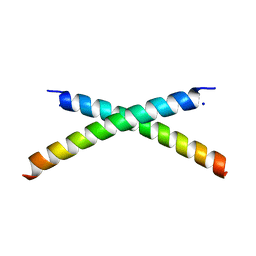 | | CRYSTAL STRUCTURE OF HUMAN PARATHYROID HORMONE 1-34 AT 0.9 A RESOLUTION | | Descriptor: | PARATHYROID HORMONE, SODIUM ION | | Authors: | Jin, L, Briggs, S.L, Chandrasekhar, S, Chirgadze, N.Y, Clawson, D.K, Schevitz, R.W, Smiley, D.L, Tashjian, A.H, Zhang, F. | | Deposit date: | 2000-04-12 | | Release date: | 2000-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Crystal structure of human parathyroid hormone 1-34 at 0.9-A resolution.
J.Biol.Chem., 275, 2000
|
|
1EZZ
 
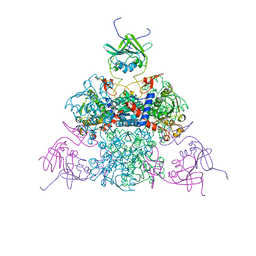 | |
3CMO
 
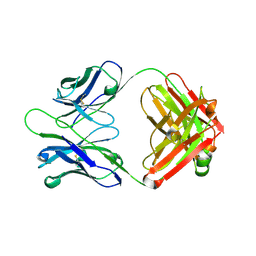 | |
3GLR
 
 | | Crystal Structure of human SIRT3 with acetyl-lysine AceCS2 peptide | | Descriptor: | Acetyl-coenzyme A synthetase 2-like, mitochondrial, BICARBONATE ION, ... | | Authors: | Jin, L, Wei, W, Jiang, Y, Peng, H, Cai, J, Mao, C, Dai, H, Bemis, J.E, Jirousek, M.R, Milne, J.C, Westphal, C.H, Perni, R.B. | | Deposit date: | 2009-03-12 | | Release date: | 2009-06-16 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Human SIRT3 Displaying Substrate-induced Conformational Changes.
J.Biol.Chem., 284, 2009
|
|
3GLT
 
 | | Crystal Structure of Human SIRT3 with ADPR bound to the AceCS2 peptide containing a thioacetyl lysine | | Descriptor: | Acetyl-coenzyme A synthetase 2-like, mitochondrial, CARBONATE ION, ... | | Authors: | Jin, L, Wei, W, Jiang, Y, Peng, H, Cai, J, Mao, C, Dai, H, Bemis, J.E, Jirousek, M.R, Milne, J.C, Westphal, C.H, Perni, R.B. | | Deposit date: | 2009-03-12 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Human SIRT3 Displaying Substrate-induced Conformational Changes.
J.Biol.Chem., 284, 2009
|
|
3GLU
 
 | | Crystal Structure of Human SIRT3 with AceCS2 peptide | | Descriptor: | Acetyl-coenzyme A synthetase 2-like, mitochondrial, NAD-dependent deacetylase sirtuin-3, ... | | Authors: | Jin, L, Wei, W, Jiang, Y, Peng, H, Cai, J, Mao, C, Dai, H, Bemis, J.E, Jirousek, M.R, Milne, J.C, Westphal, C.H, Perni, R.B. | | Deposit date: | 2009-03-12 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of Human SIRT3 Displaying Substrate-induced Conformational Changes.
J.Biol.Chem., 284, 2009
|
|
3GLS
 
 | | Crystal Structure of Human SIRT3 | | Descriptor: | NAD-dependent deacetylase sirtuin-3, mitochondrial, SULFATE ION, ... | | Authors: | Jin, L, Wei, W, Jiang, Y, Peng, H, Cai, J, Mao, C, Dai, H, Bemis, J.E, Jirousek, M.R, Milne, J.C, Westphal, C.H, Perni, R.B. | | Deposit date: | 2009-03-12 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structures of Human SIRT3 Displaying Substrate-induced Conformational Changes.
J.Biol.Chem., 284, 2009
|
|
1MF8
 
 | |
1XXF
 
 | | Crystal Structure of the FXIa Catalytic Domain in Complex with Ecotin Mutant (EcotinP) | | Descriptor: | Coagulation factor XI, Ecotin, SODIUM ION | | Authors: | Jin, L, Pandey, P, Babine, R.E, Gorga, J.C, Seidl, K.J, Gelfand, E, Weaver, D.T, Abdel-Meguid, S.S, Strickler, J.E. | | Deposit date: | 2004-11-04 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of the FXIa Catalytic Domain in Complex with Ecotin Mutants Reveal Substrate-like Interactions
J.Biol.Chem., 280, 2005
|
|
1ZHR
 
 | | Crystal Structure of the Catalytic Domain of Coagulation Factor XI in Complex with Benzamidine (S434A-T475A-C482S-K437A Mutant) | | Descriptor: | BENZAMIDINE, coagulation factor XI | | Authors: | Jin, L, Pandey, P, Babine, R.E, Weaver, D.T, Abdel-Meguid, S.S, Strickler, J.E. | | Deposit date: | 2005-04-26 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Mutation of surface residues to promote crystallization of activated factor XI as a complex with benzamidine: an essential step for the iterative structure-based design of factor XI inhibitors.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1XX9
 
 | | Crystal Structure of the FXIa Catalytic Domain in Complex with EcotinM84R | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor XI, Ecotin | | Authors: | Jin, L, Pandey, P, Babine, R.E, Gorga, J.C, Seidl, K.J, Gelfand, E, Weaver, D.T, Abdel-Meguid, S.S, Strickler, J.E. | | Deposit date: | 2004-11-04 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of the FXIa Catalytic Domain in Complex with Ecotin Mutants Reveal Substrate-like Interactions
J.Biol.Chem., 280, 2005
|
|
1XXD
 
 | | Crystal Structure of the FXIa Catalytic Domain in Complex with mutated Ecotin | | Descriptor: | Coagulation factor XI, Ecotin | | Authors: | Jin, L, Pandey, P, Babine, R.E, Gorga, J.C, Seidl, K.J, Gelfand, E, Weaver, D.T, Abdel-Meguid, S.S, Strickler, J.E. | | Deposit date: | 2004-11-04 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Crystal Structures of the FXIa Catalytic Domain in Complex with Ecotin Mutants Reveal Substrate-like Interactions
J.Biol.Chem., 280, 2005
|
|
1ZHM
 
 | | Crystal Structure of the Catalytic Domain of the Coagulation Factor XIa in Complex with Benzamidine (S434A-T475A-K437 Mutant) | | Descriptor: | BENZAMIDINE, GLUTATHIONE, coagulation factor XI | | Authors: | Jin, L, Pandey, P, Babine, R.E, Weaver, D.T, Abdel-Meguid, S.S, Strickler, J.E. | | Deposit date: | 2005-04-26 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Mutation of surface residues to promote crystallization of activated factor XI as a complex with benzamidine: an essential step for the iterative structure-based design of factor XI inhibitors.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1ZHP
 
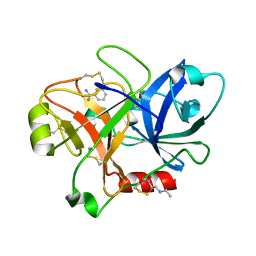 | | Crystal Structure of the Catalytic Domain of Coagulation Factor XI in Complex with Benzamidine (S434A-T475A-K505 Mutant) | | Descriptor: | BENZAMIDINE, GLUTATHIONE, coagulation factor XI | | Authors: | Jin, L, Pandey, P, Babine, R.E, Weaver, D.T, Abdel-Meguid, S.S, Strickler, J.E. | | Deposit date: | 2005-04-26 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mutation of surface residues to promote crystallization of activated factor XI as a complex with benzamidine: an essential step for the iterative structure-based design of factor XI inhibitors.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1KHY
 
 | |
7RWG
 
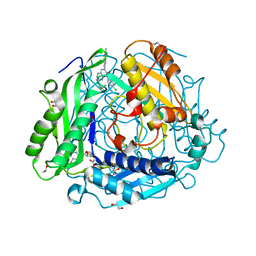 | | "Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric inhibitor AGI-43192 | | Descriptor: | (8R)-8-(4-chlorophenyl)-6-(2-methyl-2H-indazol-5-yl)-2-[(2,2,2-trifluoroethyl)amino]-5,8-dihydropyrido[4,3-d]pyrimidin-7(6H)-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Jin, L, Padyana, A.K. | | Deposit date: | 2021-08-19 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Leveraging Structure-Based Drug Design to Identify Next-Generation MAT2A Inhibitors, Including Brain-Penetrant and Peripherally Efficacious Leads.
J.Med.Chem., 65, 2022
|
|
7RWH
 
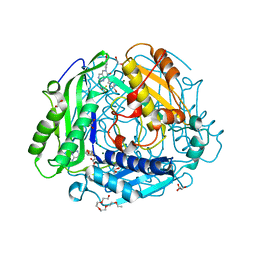 | | Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric inhibitor AGI-41998 | | Descriptor: | 1,2-ETHANEDIOL, 8-(4-bromophenyl)-6-(4-methoxyphenyl)-2-[2,2,2-tris(fluoranyl)ethylamino]pyrido[4,3-d]pyrimidin-7-ol, CHLORIDE ION, ... | | Authors: | Jin, L, Padyana, A.K. | | Deposit date: | 2021-08-19 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Leveraging Structure-Based Drug Design to Identify Next-Generation MAT2A Inhibitors, Including Brain-Penetrant and Peripherally Efficacious Leads.
J.Med.Chem., 65, 2022
|
|
7RW7
 
 | | Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric inhibitor Compound 9 | | Descriptor: | (3'R)-2-[(cyclopropylmethyl)amino]-6-(4-methoxyphenyl)-1'-[(1H-pyrazol-5-yl)methyl]-5,6-dihydro-7H-spiro[pyrido[4,3-d]pyrimidine-8,3'-pyrrolidin]-7-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Jin, L, Padyana, A.K. | | Deposit date: | 2021-08-19 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Leveraging Structure-Based Drug Design to Identify Next-Generation MAT2A Inhibitors, Including Brain-Penetrant and Peripherally Efficacious Leads.
J.Med.Chem., 65, 2022
|
|
7RW5
 
 | | Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric inhibitor Compound 1 | | Descriptor: | (3'R)-N-(cyclopropylmethyl)-1'-[(2-fluorophenyl)methyl]-4-methyl-5H,7H-spiro[pyrano[4,3-d]pyrimidine-8,3'-pyrrolidin]-2-amine, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | Authors: | Jin, L, Padyana, A.K. | | Deposit date: | 2021-08-19 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Leveraging Structure-Based Drug Design to Identify Next-Generation MAT2A Inhibitors, Including Brain-Penetrant and Peripherally Efficacious Leads.
J.Med.Chem., 65, 2022
|
|
1AKM
 
 | |
