1LLD
 
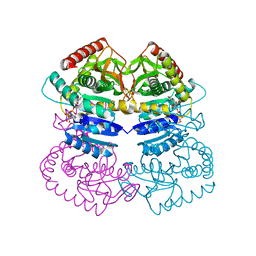 | |
1LTH
 
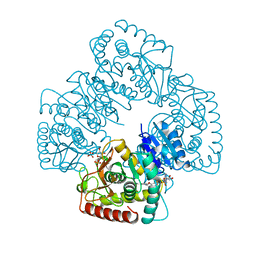 | |
1BGY
 
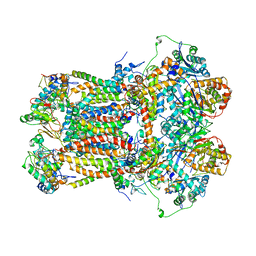 | | CYTOCHROME BC1 COMPLEX FROM BOVINE | | Descriptor: | CYTOCHROME BC1 COMPLEX, FE2/S2 (INORGANIC) CLUSTER, HEME C, ... | | Authors: | Iwata, S, Lee, J.W, Okada, K, Lee, J.K, Iwata, M, Ramaswamy, S, Jap, B.K. | | Deposit date: | 1998-06-02 | | Release date: | 1999-01-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex.
Science, 281, 1998
|
|
1BE3
 
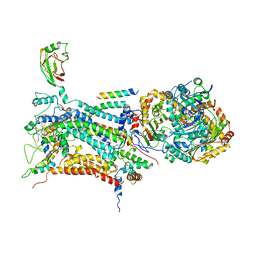 | | CYTOCHROME BC1 COMPLEX FROM BOVINE | | Descriptor: | CYTOCHROME BC1 COMPLEX, FE2/S2 (INORGANIC) CLUSTER, HEME C, ... | | Authors: | Iwata, S, Lee, J.W, Okada, K, Lee, J.K, Iwata, M, Ramaswamy, S, Jap, B.K. | | Deposit date: | 1998-05-19 | | Release date: | 1999-01-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex.
Science, 281, 1998
|
|
1RIE
 
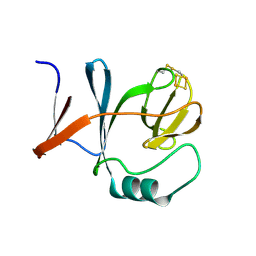 | | STRUCTURE OF A WATER SOLUBLE FRAGMENT OF THE RIESKE IRON-SULFUR PROTEIN OF THE BOVINE HEART MITOCHONDRIAL CYTOCHROME BC1-COMPLEX | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, RIESKE IRON-SULFUR PROTEIN | | Authors: | Iwata, S, Saynovits, M, Link, T.A, Michel, H. | | Deposit date: | 1996-02-23 | | Release date: | 1996-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a water soluble fragment of the 'Rieske' iron-sulfur protein of the bovine heart mitochondrial cytochrome bc1 complex determined by MAD phasing at 1.5 A resolution.
Structure, 4, 1996
|
|
7C4S
 
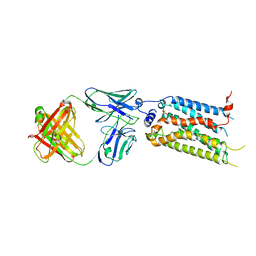 | | Sphingosine-1-phosphate receptor 3 with a natural ligand. | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Antibody Fab fragment heavy chain, Antibody Fab fragment light chain, ... | | Authors: | Iwata, S, Maeda, S, Luo, F, Nango, E, hirata, K, Asada, H. | | Deposit date: | 2020-05-18 | | Release date: | 2021-06-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Endogenous agonist-bound S1PR3 structure reveals determinants of G protein-subtype bias.
Sci Adv, 7, 2021
|
|
8ZLR
 
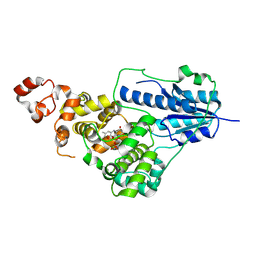 | | SFX structure of CraCRY in the semiquinone state | | Descriptor: | Cryptochrome photoreceptor, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE | | Authors: | Maestre-Reyna, M, Hosokawa, Y, Wang, P.-H, Saft, M, Caramello, N, Engilberge, S, Franz-Badur, S, Ngura Putu, E.P.G, Nakamura, M, Wu, W.-J, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Yang, C.-H, Lin, W.-T, Yang, K.-C, Ban, Y, Imura, T, Kazuoka, A, Tanida, E, Owada, S, Joti, Y, Tanaka, R, Tanaka, T, Luo, F, Tono, K, Kiontke, S, Korf, L, Umena, Y, Tosha, T, Bessho, Y, Nango, E, Iwata, S, Royant, A, Tsai, M.-D, Yamamoto, J, Essen, L.-O. | | Deposit date: | 2024-05-21 | | Release date: | 2025-05-14 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Capturing structural intermediates in an animal-like cryptochrome photoreceptor by time-resolved crystallography.
Sci Adv, 11, 2025
|
|
3GQB
 
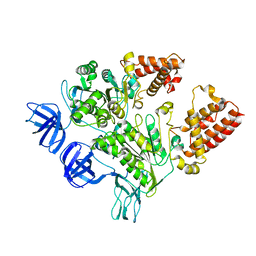 | | Crystal Structure of the A3B3 complex from V-ATPase | | Descriptor: | V-type ATP synthase alpha chain, V-type ATP synthase beta chain | | Authors: | Meher, M, Akimoto, S, Iwata, M, Nagata, K, Hori, Y, Yoshida, M, Yokoyama, S, Iwata, S, Yokoyama, K. | | Deposit date: | 2009-03-24 | | Release date: | 2009-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of A(3)B(3) complex of V-ATPase from Thermus thermophilus.
Embo J., 28, 2009
|
|
1KQG
 
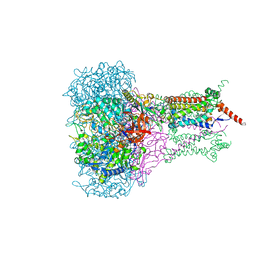 | | FORMATE DEHYDROGENASE N FROM E. COLI | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, 2-HEPTYL-4-HYDROXY QUINOLINE N-OXIDE, CARDIOLIPIN, ... | | Authors: | Jormakka, M, Tornroth, S, Byrne, B, Iwata, S. | | Deposit date: | 2002-01-05 | | Release date: | 2002-03-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis of proton motive force generation: structure of formate dehydrogenase-N.
Science, 295, 2002
|
|
2V8N
 
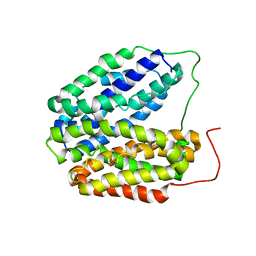 | | Wild-type Structure of Lactose Permease | | Descriptor: | LACTOSE PERMEASE | | Authors: | Guan, L, Mirza, O, Verner, G, Iwata, S, Kaback, H.R. | | Deposit date: | 2007-08-09 | | Release date: | 2007-09-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural Determination of Wild-Type Lactose Permease.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
4W4Q
 
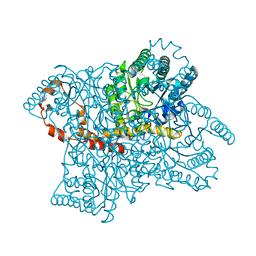 | | Glucose isomerase structure determined by serial femtosecond crystallography at SACLA | | Descriptor: | CALCIUM ION, Xylose isomerase | | Authors: | Nango, E, Tanaka, T, Sugahara, M, Suzuki, M, Iwata, S. | | Deposit date: | 2014-08-15 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Grease matrix as a versatile carrier of proteins for serial crystallography
Nat.Methods, 12, 2015
|
|
1FFT
 
 | | The structure of ubiquinol oxidase from Escherichia coli | | Descriptor: | COPPER (II) ION, HEME O, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Abramson, J, Riistama, S, Larsson, G, Jasaitis, A, Svensson-Ek, M, Puustinen, A, Iwata, S, Wikstrom, M. | | Deposit date: | 2000-07-26 | | Release date: | 2000-10-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structure of the ubiquinol oxidase from Escherichia coli and its ubiquinone binding site.
Nat.Struct.Biol., 7, 2000
|
|
2D00
 
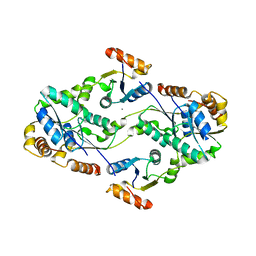 | | Subunit F of V-type ATPase/synthase | | Descriptor: | CALCIUM ION, V-type ATP synthase subunit F | | Authors: | Makyio, H, Iino, R, Ikeda, C, Imamura, H, Tamakoshi, M, Iwata, M, Stock, D, Bernal, R.A, Carpenter, E.P, Yoshida, M, Yokoyama, K, Iwata, S. | | Deposit date: | 2005-07-21 | | Release date: | 2005-12-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a central stalk subunit F of prokaryotic V-type ATPase/synthase from Thermus thermophilus
Embo J., 24, 2005
|
|
4V4O
 
 | | Crystal Structure of the Chaperonin Complex Cpn60/Cpn10/(ADP)7 from Thermus Thermophilus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Shimamura, T, Koike-Takeshita, A, Yokoyama, K, Masui, R, Murai, N, Yoshida, M, Taguchi, H, Iwata, S. | | Deposit date: | 2004-05-23 | | Release date: | 2014-07-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the native chaperonin complex from Thermus thermophilus revealed unexpected asymmetry at the cis-cavity
STRUCTURE, 12, 2004
|
|
1R5Z
 
 | | Crystal Structure of Subunit C of V-ATPase | | Descriptor: | V-type ATP synthase subunit C | | Authors: | Iwata, M, Imamura, H, Stambouli, E, Ikeda, C, Tamakoshi, M, Nagata, K, Makyio, H, Hankamer, B, Barber, J, Yoshida, M, Yokoyama, K, Iwata, S. | | Deposit date: | 2003-10-14 | | Release date: | 2004-01-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of a central stalk subunit C and reversible association/dissociation of vacuole-type ATPase.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
7F8T
 
 | | Re-refinement of the 2XRY X-ray structure of archaeal class II CPD photolyase from Methanosarcina mazei | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.C, Nango, E, Gusti-Ngurah-Putu, E.-P, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Sugahara, M, Owada, S, Joti, Y, Tanaka, R, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-07-02 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
5XJM
 
 | | Complex structure of angiotensin II type 2 receptor with Fab | | Descriptor: | FabH, FabL, Sar1, ... | | Authors: | Asada, H, Horita, S, Shimamura, T, Iwata, S. | | Deposit date: | 2017-05-02 | | Release date: | 2018-07-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the human angiotensin II type 2 receptor bound to an angiotensin II analog
Nat. Struct. Mol. Biol., 25, 2018
|
|
5XLI
 
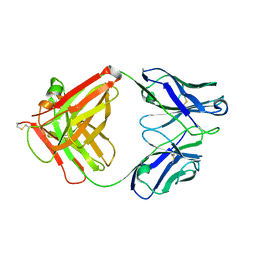 | | Structure of anti-Angiotensin II type2 receptor antibody (D5711-4A03) | | Descriptor: | FabH, FabL | | Authors: | Asada, H, Horita, S, Iwata, S, Hirata, K. | | Deposit date: | 2017-05-10 | | Release date: | 2018-07-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Crystal structure of the human angiotensin II type 2 receptor bound to an angiotensin II analog.
Nat. Struct. Mol. Biol., 25, 2018
|
|
4GAV
 
 | | Structure of the Ndi1 protein from Saccharomyces cerevisiae in complex with quinone | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Rotenone-insensitive NADH-ubiquinone oxidoreductase, UBIQUINONE-2 | | Authors: | Iwata, M, Lee, Y, Yamashita, T, Yagi, T, Iwata, S, Cameron, A.D, Maher, M.J. | | Deposit date: | 2012-07-25 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of the yeast NADH dehydrogenase (Ndi1) reveals overlapping binding sites for water- and lipid-soluble substrates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4G9K
 
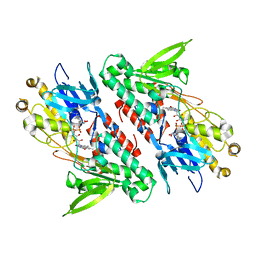 | | Structure of the Ndi1 protein from Saccharomyces cerevisiae | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Rotenone-insensitive NADH-ubiquinone oxidoreductase | | Authors: | Iwata, M, Lee, Y, Yamashita, T, Yagi, T, Iwata, S, Cameron, A.D, Maher, M.J. | | Deposit date: | 2012-07-24 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of the yeast NADH dehydrogenase (Ndi1) reveals overlapping binding sites for water- and lipid-soluble substrates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4GAP
 
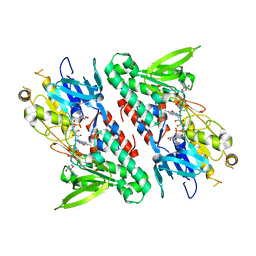 | | Structure of the Ndi1 protein from Saccharomyces cerevisiae in complex with NAD+ | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Rotenone-insensitive NADH-ubiquinone oxidoreductase | | Authors: | Iwata, M, Lee, Y, Yamashita, T, Yagi, T, Iwata, S, Cameron, A.D, Maher, M.J. | | Deposit date: | 2012-07-25 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of the yeast NADH dehydrogenase (Ndi1) reveals overlapping binding sites for water- and lipid-soluble substrates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4D1C
 
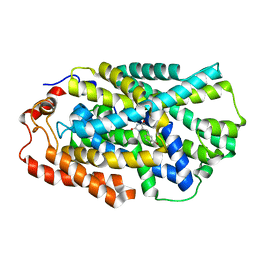 | | STRUCTURE OF MHP1, A NUCLEOBASE-CATION-SYMPORT-1 FAMILY TRANSPORTER, IN A CLOSED CONFORMATION WITH bromovinylhydantoin bound. | | Descriptor: | (5Z)-5-[(3-bromophenyl)methylidene]imidazolidine-2,4-dione, HYDANTOIN TRANSPORT PROTEIN, SODIUM ION | | Authors: | Weyand, S, Brueckner, F, Geng, T, Drew, D, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Molecular Mechanism of Ligand Recognition by Membrane Transport Protein, Mhp1.
Embo J., 33, 2014
|
|
4D1D
 
 | | STRUCTURE OF MHP1, A NUCLEOBASE-CATION-SYMPORT-1 FAMILY TRANSPORTER with the inhibitor 5-(2-naphthylmethyl)-L-hydantoin. | | Descriptor: | 5-(2-NAPHTHYLMETHYL)-D-HYDANTOIN, 5-(2-NAPHTHYLMETHYL)-L-HYDANTOIN, HYDANTOIN TRANSPORT PROTEIN, ... | | Authors: | Weyand, S, Brueckner, F, Geng, T, Drew, D, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Molecular Mechanism of Ligand Recognition by Membrane Transport Protein, Mhp1.
Embo J., 33, 2014
|
|
4D1B
 
 | | STRUCTURE OF MHP1, A NUCLEOBASE-CATION-SYMPORT-1 FAMILY TRANSPORTER, IN A CLOSED CONFORMATION WITH BENZYL-HYDANTOIN | | Descriptor: | (5S)-5-benzylimidazolidine-2,4-dione, HYDANTOIN TRANSPORT PROTEIN, SODIUM ION | | Authors: | Brueckner, F, Geng, T, Weyand, S, Drew, D, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Molecular Mechanism of Ligand Recognition by Membrane Transport Protein, Mhp1.
Embo J., 33, 2014
|
|
4D1A
 
 | | STRUCTURE OF MHP1, A NUCLEOBASE-CATION-SYMPORT-1 FAMILY TRANSPORTER, IN A CLOSED CONFORMATION WITH INDOLYLMETHYL-HYDANTOIN | | Descriptor: | (5S)-5-(1H-indol-3-ylmethyl)imidazolidine-2,4-dione, HYDANTOIN TRANSPORT PROTEIN, SODIUM ION | | Authors: | Weyand, S, Brueckner, F, Geng, T, Drew, D, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Molecular Mechanism of Ligand Recognition by Membrane Transport Protein, Mhp1.
Embo J., 33, 2014
|
|
