7CG3
 
 | | Staggered ring conformation of CtHsp104 (Hsp104 from Chaetomium Thermophilum) | | Descriptor: | Heat shock protein 104 | | Authors: | Inoue, Y, Hanazono, Y, Noi, K, Kawamoto, A, Kimatsuka, M, Harada, R, Takeda, K, Iwamasa, N, Shibata, K, Noguchi, K, Shigeta, Y, Namba, K, Ogura, T, Miki, K, Shinohara, K, Yohda, M. | | Deposit date: | 2020-06-30 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Split conformation of Chaetomium thermophilum Hsp104 disaggregase.
Structure, 29, 2021
|
|
7VR5
 
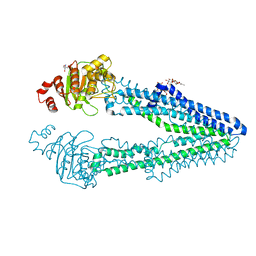 | | Crystal structure of CmABCB1 W114Y/W161Y/W363Y/W364Y/M391W (4WY/M391W) mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DECYL-BETA-D-MALTOPYRANOSIDE, Probable ATP-dependent transporter ycf16 | | Authors: | Inoue, Y, Ogawa, H, Kato, H. | | Deposit date: | 2021-10-21 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-based alteration of tryptophan residues of the multidrug transporter CmABCB1 to assess substrate binding using fluorescence spectroscopy.
Protein Sci., 31, 2022
|
|
5ZUI
 
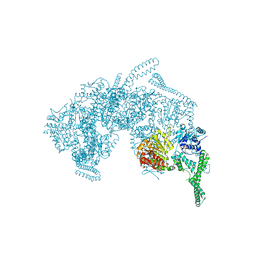 | | Crystal Structure of HSP104 from Chaetomium thermophilum | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Heat Shock Protein 104, SULFATE ION | | Authors: | Hanazono, Y, Inoue, Y, Noguchi, K, Yohda, M, Shinohara, K, Takeda, K, Miki, K. | | Deposit date: | 2018-05-07 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Split conformation of Chaetomium thermophilum Hsp104 disaggregase.
Structure, 2021
|
|
7CTN
 
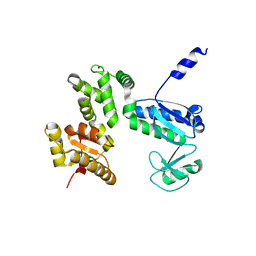 | | Structure of the 328-692 fragment of FlhA (E351A/D356A) | | Descriptor: | Flagellar biosynthesis protein FlhA | | Authors: | Kida, M, Takekawa, N, Kinoshita, M, Inoue, Y, Minamino, T, Imada, K. | | Deposit date: | 2020-08-19 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The FlhA linker mediates flagellar protein export switching during flagellar assembly.
Commun Biol, 4, 2021
|
|
1SPA
 
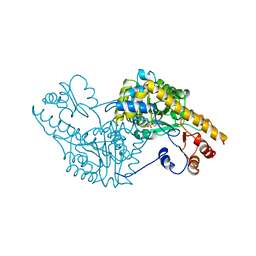 | | ROLE OF ASP222 IN THE CATALYTIC MECHANISM OF ESCHERICHIA COLI ASPARTATE AMINOTRANSFERASE: THE AMINO ACID RESIDUE WHICH ENHANCES THE FUNCTION OF THE ENZYME-BOUND COENZYME PYRIDOXAL 5'-PHOSPHATE | | Descriptor: | ASPARTATE AMINOTRANSFERASE, N-METHYL-4-DEOXY-4-AMINO-PYRIDOXAL-5-PHOSPHATE | | Authors: | Hinoue, Y, Yano, T, Metzler, D.E, Miyahara, I, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 1993-01-26 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of Asp222 in the catalytic mechanism of Escherichia coli aspartate aminotransferase: the amino acid residue which enhances the function of the enzyme-bound coenzyme pyridoxal 5'-phosphate.
Biochemistry, 31, 1992
|
|
1AB9
 
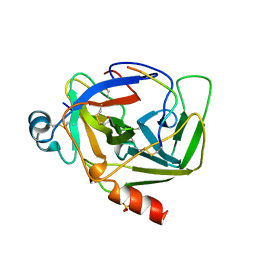 | | CRYSTAL STRUCTURE OF BOVINE GAMMA-CHYMOTRYPSIN | | Descriptor: | GAMMA-CHYMOTRYPSIN, PENTAPEPTIDE (TPGVY), SULFATE ION | | Authors: | Sugio, S, Kashima, A, Inoue, Y, Maeda, I, Nose, T, Shimohigashi, Y. | | Deposit date: | 1997-02-05 | | Release date: | 1997-08-20 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystal structure of a dipeptide-chymotrypsin complex in an inhibitory interaction.
Eur.J.Biochem., 255, 1998
|
|
1AFQ
 
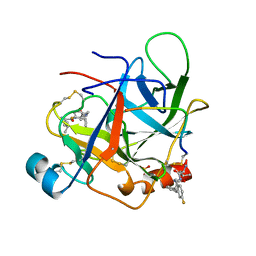 | | CRYSTAL STRUCTURE OF BOVINE GAMMA-CHYMOTRYPSIN COMPLEXED WITH A SYNTHETIC INHIBITOR | | Descriptor: | BOVINE GAMMA-CHYMOTRYPSIN, D-leucyl-N-(4-fluorobenzyl)-L-phenylalaninamide, SULFATE ION | | Authors: | Sugio, S, Kashima, A, Inoue, Y, Maeda, I, Nose, T, Shimohigashi, Y. | | Deposit date: | 1997-03-12 | | Release date: | 1997-09-17 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of a dipeptide-chymotrypsin complex in an inhibitory interaction.
Eur.J.Biochem., 255, 1998
|
|
1EE8
 
 | | CRYSTAL STRUCTURE OF MUTM (FPG) PROTEIN FROM THERMUS THERMOPHILUS HB8 | | Descriptor: | MUTM (FPG) PROTEIN, ZINC ION | | Authors: | Sugahara, M, Mikawa, T, Kumasaka, T, Yamamoto, M, Kato, R, Fukuyama, K, Inoue, Y, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2000-01-31 | | Release date: | 2001-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a repair enzyme of oxidatively damaged DNA, MutM (Fpg), from an extreme thermophile, Thermus thermophilus HB8.
EMBO J., 19, 2000
|
|
8KGA
 
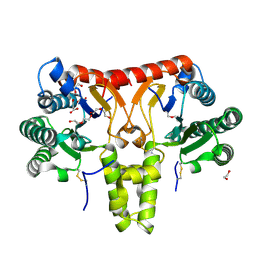 | | SlNDPS1-AtcPT4 Chimera | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Dimethylallylcistransferase CPT1, ... | | Authors: | Suenaga-Hiromori, M, Ishii, T, Imaizumi, R, Takeshita, K, Yanai, T, Matsuura, H, Sakai, N, Yamaguchi, H, Yanbe, F, Waki, T, Tozawa, Y, Miyagi-Inoue, Y, Yamamoto, M, Kataoka, K, Nakayama, T, Yamashita, S, Takahashi, S. | | Deposit date: | 2023-08-18 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.182 Å) | | Cite: | Biosynthesising various rubber-like polymers by reconstituting prenyltransferases on Hevea rubber particles: molecular and structural bases of the versatile system
To Be Published
|
|
8KGB
 
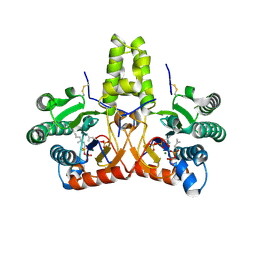 | | SlNDPS1-AtcPT4 Chimera complexed with GSPP, Mg2+, and IPP | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Dimethylallylcistransferase CPT1, chloroplastic,Dehydrodolichyl diphosphate synthase 2, ... | | Authors: | Suenaga-Hiromori, M, Ishii, T, Imaizumi, R, Takeshita, K, Yanai, T, Matsuura, H, Sakai, N, Yamaguchi, H, Yanbe, F, Waki, T, Tozawa, Y, Miyagi-Inoue, Y, Yamamoto, M, Kataoka, K, Nakayama, T, Yamashita, S, Takahashi, S. | | Deposit date: | 2023-08-18 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A versatile system for enzymatic synthesis of natural and unnatural polyisoprenoids on rubber particles
To Be Published
|
|
7V2G
 
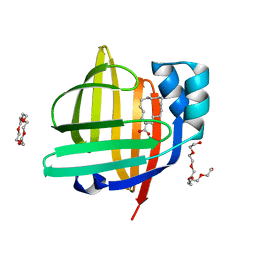 | | The 0.98 angstrom structure of the human FABP3 Y19F mutant complexed with palmitic acid | | Descriptor: | Fatty acid-binding protein, heart, HEXAETHYLENE GLYCOL, ... | | Authors: | Sugiyama, S, Takahashi, J, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-08-09 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | The 0.98 angstrom structure of the human FABP3 Y19F mutant complexed with palmitic acid
To Be Published
|
|
1GD7
 
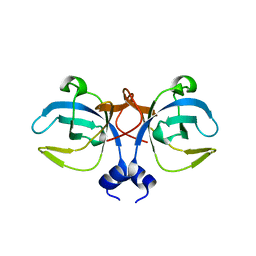 | | CRYSTAL STRUCTURE OF A BIFUNCTIONAL PROTEIN (CSAA) WITH EXPORT-RELATED CHAPERONE AND TRNA-BINDING ACTIVITIES. | | Descriptor: | CSAA PROTEIN | | Authors: | Shibata, T, Inoue, Y, Vassylyev, D.G, Kawaguchi, S, Yokoyama, S, Muller, J, Linde, D, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2000-09-22 | | Release date: | 2001-09-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the ttCsaA protein: an export-related chaperone from Thermus thermophilus.
EMBO J., 20, 2001
|
|
2RQ9
 
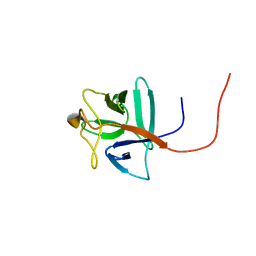 | | Solution structure of human acidic fibroblast growth factor (aFGF) in the presence of a protein stabilizer NDSB-new | | Descriptor: | Heparin-binding growth factor 1 | | Authors: | Enomoto, M, Xiang, L, Ishii, T, Tochio, N, Hosoda, K, Inoue, Y, Nameki, N, Wakamatsu, K. | | Deposit date: | 2009-03-12 | | Release date: | 2010-03-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human acidic fibroblast growth factor (aFGF) in the presence of a protein stabilizer NDSB-new
To be Published
|
|
8X36
 
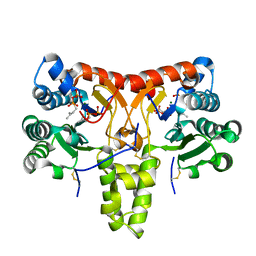 | | Neryl diphosphate synthase from Solanum lycopersicum complexed with DMSAPP, IPP, and magnesium ion (form B) | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, DIMETHYLALLYL S-THIOLODIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Imaizumi, R, Matsuura, H, Yanai, T, Takeshita, K, Misawa, S, Yamaguchi, H, Sakai, N, Miyagi-Inoue, Y, Suenaga-Hiromori, M, Kataoka, K, Nakayama, T, Yamamoto, M, Takahashi, S, Yamashita, S. | | Deposit date: | 2023-11-12 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural-Functional Correlations between Unique N-terminal Region and C-terminal Conserved Motif in Short-chain cis-Prenyltransferase from Tomato.
Chembiochem, 25, 2024
|
|
8X35
 
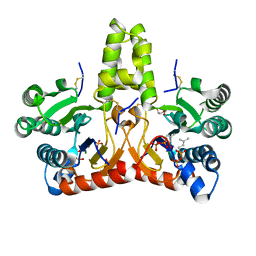 | | Neryl diphosphate synthase from Solanum lycopersicum complexed with DMSAPP, IPP, and magnesium ion (form A) | | Descriptor: | 1,2-ETHANEDIOL, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Imaizumi, R, Matsuura, H, Yanai, T, Takeshita, K, Misawa, S, Yamaguchi, H, Sakai, N, Miyagi-Inoue, Y, Suenaga-Hiromori, M, Kataoka, K, Nakayama, T, Yamamoto, M, Takahashi, S, Yamashita, S. | | Deposit date: | 2023-11-12 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural-Functional Correlations between Unique N-terminal Region and C-terminal Conserved Motif in Short-chain cis-Prenyltransferase from Tomato.
Chembiochem, 25, 2024
|
|
8X37
 
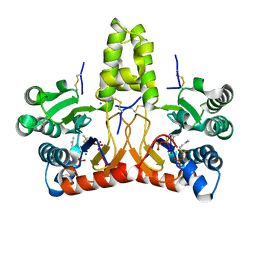 | | Neryl diphosphate synthase from Solanum lycopersicum complexed with DMSAPP | | Descriptor: | DIMETHYLALLYL S-THIOLODIPHOSPHATE, MAGNESIUM ION, Neryl-diphosphate synthase 1 | | Authors: | Imaizumi, R, Matsuura, H, Yanai, T, Takeshita, K, Misawa, S, Yamaguchi, H, Sakai, N, Miyagi-Inoue, Y, Suenaga-Hiromori, M, Kataoka, K, Nakayama, T, Yamamoto, M, Takahashi, S, Yamashita, S. | | Deposit date: | 2023-11-12 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural-Functional Correlations between Unique N-terminal Region and C-terminal Conserved Motif in Short-chain cis-Prenyltransferase from Tomato.
Chembiochem, 25, 2024
|
|
5GVO
 
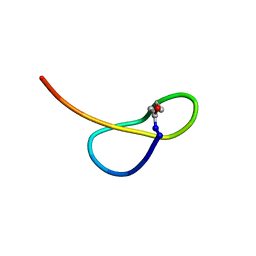 | | Solution NMR structure of a new lasso peptide sphaericin | | Descriptor: | Uncharacterized protein | | Authors: | Hemmi, H, Kodani, S, Inoue, Y, Suzuki, M, Dohra, H, Suzuki, T, Ohnishi-Kameyama, M. | | Deposit date: | 2016-09-06 | | Release date: | 2017-04-05 | | Method: | SOLUTION NMR | | Cite: | Sphaericin, a Lasso Peptide from the Rare Actinomycete Planomonospora sphaerica
Eur.J.Org.Chem., 2017
|
|
7EGO
 
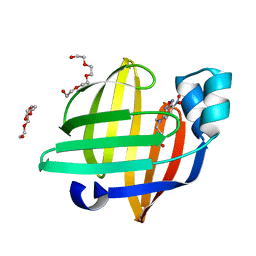 | | X-ray structure of the human heart fatty acid-binding protein complexed with the fluorescent probe HA527 | | Descriptor: | 3-[methyl-(4-nitro-2,1,3-benzoxadiazol-7-yl)amino]propanoic acid, Fatty acid-binding protein, heart, ... | | Authors: | Takabayashi, M, Yokota, J, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Sugiyama, S. | | Deposit date: | 2021-03-24 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | X-ray structure of the human heart fatty acid-binding protein complexed with the fluorescent probe HA527
To Be Published
|
|
1IPA
 
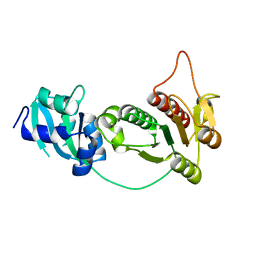 | | CRYSTAL STRUCTURE OF RNA 2'-O RIBOSE METHYLTRANSFERASE | | Descriptor: | RNA 2'-O-RIBOSE METHYLTRANSFERASE | | Authors: | Nureki, O, Shirouzu, M, Hashimoto, K, Ishitani, R, Terada, T, Tamakoshi, M, Oshima, T, Chijimatsu, M, Takio, K, Vassylyev, D.G, Shibata, T, Inoue, Y, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-05-02 | | Release date: | 2002-07-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An enzyme with a deep trefoil knot for the active-site architecture.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1IQU
 
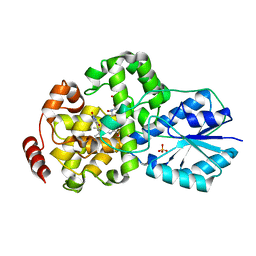 | | Crystal structure of photolyase-thymine complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION, THYMINE, ... | | Authors: | Komori, H, Masui, R, Kuramitsu, S, Yokoyama, S, Shibata, T, Inoue, Y, Miki, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-08-03 | | Release date: | 2002-05-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of thermostable DNA photolyase: pyrimidine-dimer recognition mechanism.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1J1H
 
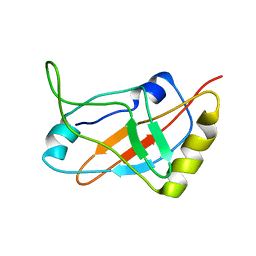 | | Solution structure of a tmRNA-binding protein, SmpB, from Thermus thermophilus | | Descriptor: | Small Protein B | | Authors: | Someya, T, Nameki, N, Hosoi, H, Suzuki, S, Hatanaka, H, Fujii, M, Terada, T, Shirouzu, M, Inoue, Y, Shibata, T, Kuramitsu, S, Yokoyama, S, Kawai, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-12-04 | | Release date: | 2003-02-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a tmRNA-binding protein, SmpB, from Thermus thermophilus
FEBS Lett., 535, 2003
|
|
7EUW
 
 | | X-ray structure of high-strength hydrogel-grown FABP3 crystal soaked in 50% DMSO solution containing 4-[2-[1-(4-bromophenyl)-5-phenyl-1H-pyrazol-3-yl]phenoxy] (HA174) | | Descriptor: | 4-[2-[1-(4-bromophenyl)-5-phenyl-pyrazol-3-yl]phenoxy]butanoic acid, Fatty acid-binding protein, heart, ... | | Authors: | Sugiyama, S, Kakinouchi, K, Nakano, R, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-05-19 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray structure of the human heart fatty acid-binding protein complexed with 4-[2-[1-(4-bromophenyl)-5-phenyl-1H-pyrazol-3-yl]phenoxy] (HA176)
To Be Published
|
|
7EUV
 
 | | X-ray structure of high-strength hydrogel-grown FABP3 crystal soaked in 50% DMSO solution containing Flurbiprofen | | Descriptor: | (2R)-2-(3-fluoro-4-phenyl-phenyl)propanoic acid, Fatty acid-binding protein, heart | | Authors: | Sugiyama, S, Kakinouchi, K, Hoshina, M, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-05-19 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | X-ray structure of the human heart fatty acid-binding protein complexed with Flurbiprofen
To Be Published
|
|
7FBF
 
 | | The 0.86 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with octanoic acid | | Descriptor: | Fatty acid-binding protein, heart, HEXAETHYLENE GLYCOL, ... | | Authors: | Sugiyama, S, Kakinouchi, K, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-07-09 | | Release date: | 2022-07-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.86 Å) | | Cite: | The 0.86 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with octanoic acid
To Be Published
|
|
7FBN
 
 | | X-ray structure of high-strength hydrogel-grown FABP3 crystal soaked in 50% DMF solution containing dibutylhydroxytoluene (BHT) | | Descriptor: | 2,6-ditert-butyl-4-methyl-phenol, ACETIC ACID, Fatty acid-binding protein, ... | | Authors: | Sugiyama, S, Kakinouchi, K, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-07-11 | | Release date: | 2022-07-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | X-ray structure of high-strength hydrogel-grown FABP3 crystal soaked in 50% DMF solution containing dibutylhydroxytoluene (BHT)
To Be Published
|
|
