7L18
 
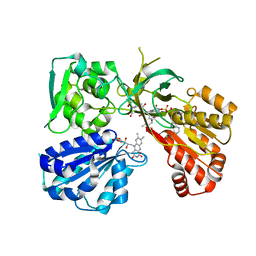 | | Crystal structure of a tandem deletion mutant of rat NADPH-cytochrome P450 reductase | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Hubbard, P.A, Xia, C, Shen, A.L, Kim, J.J.K. | | Deposit date: | 2020-12-14 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.542 Å) | | Cite: | Structural and kinetic investigations of the carboxy terminus of NADPH-cytochrome P450 oxidoreductase.
Arch.Biochem.Biophys., 701, 2021
|
|
1XX4
 
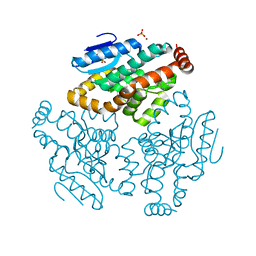 | | Crystal Structure of Rat Mitochondrial 3,2-Enoyl-CoA | | Descriptor: | 3,2-trans-enoyl-CoA isomerase, mitochondrial, BENZAMIDINE, ... | | Authors: | Hubbard, P.A, Yu, W, Schulz, H, Kim, J.-J. | | Deposit date: | 2004-11-03 | | Release date: | 2004-11-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Domain swapping in the low-similarity isomerase/hydratase superfamily: the crystal structure of rat mitochondrial Delta3, Delta2-enoyl-CoA isomerase.
Protein Sci., 14, 2005
|
|
5VY1
 
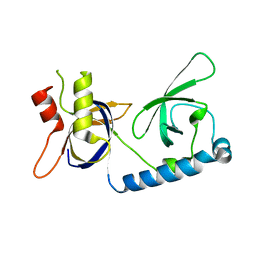 | | Crystal structure of the extended Tudor domain from BmPAPI | | Descriptor: | Tudor and KH domain-containing protein homolog | | Authors: | Hubbard, P.A, Pan, X, McNally, R, Ohtaki, A, Honda, S, Kirino, Y, Murali, R. | | Deposit date: | 2017-05-24 | | Release date: | 2017-06-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural studies of the Tudor domain from the Bombyx homolog of Drosophila PAPI: Implication to piRNA biogenesis
To Be Published
|
|
5VQH
 
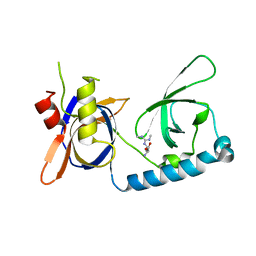 | | Crystal structure of the extended Tudor domain from BmPAPI in complex with sDMA | | Descriptor: | N3, N4-DIMETHYLARGININE, Tudor and KH domain-containing protein homolog | | Authors: | Hubbard, P.A, Pan, X, Ohtaki, A, McNally, R, Honda, S, Kirino, Y, Murali, R. | | Deposit date: | 2017-05-08 | | Release date: | 2017-06-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural studies of the Tudor domain from the Bombyx homolog of Drosophila PAPI: Implication to piRNA biogenesis
To Be Published
|
|
5VQG
 
 | | Crystal structure of the extended Tudor domain from BmPAPI | | Descriptor: | Tudor and KH domain-containing protein homolog | | Authors: | Hubbard, P.A, Pan, X, Ohtaki, A, McNally, R, Honda, S, Kirino, Y, Murali, R. | | Deposit date: | 2017-05-08 | | Release date: | 2017-06-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural studies of the Tudor domain from the Bombyx homolog of Drosophila PAPI: Implication to piRNA biogenesis
To Be Published
|
|
2QM7
 
 | | MeaB, A Bacterial Homolog of MMAA, Bound to GDP | | Descriptor: | GTPase/ATPase, GUANOSINE-5'-DIPHOSPHATE, PHOSPHATE ION | | Authors: | Hubbard, P.A, Padovani, D, Labunska, T, Mahlstedt, S.A, Banerjee, R, Drennan, C.L. | | Deposit date: | 2007-07-14 | | Release date: | 2007-08-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure and mutagenesis of the metallochaperone MeaB: insight into the causes of methylmalonic aciduria.
J.Biol.Chem., 282, 2007
|
|
2QM8
 
 | | MeaB, A Bacterial Homolog of MMAA, in the Nucleotide Free Form | | Descriptor: | GTPase/ATPase, PHOSPHATE ION | | Authors: | Hubbard, P.A, Padovani, D, Labunska, T, Mahlstedt, S.A, Banerjee, R, Drennan, C.L. | | Deposit date: | 2007-07-14 | | Release date: | 2007-08-28 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure and mutagenesis of the metallochaperone MeaB: insight into the causes of methylmalonic aciduria.
J.Biol.Chem., 282, 2007
|
|
1JA0
 
 | | CYPOR-W677X | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Hubbard, P.A, Shen, A.L, Paschke, R, Kasper, C.B, Kim, J.J. | | Deposit date: | 2001-05-29 | | Release date: | 2001-08-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | NADPH-cytochrome P450 oxidoreductase. Structural basis for hydride and electron transfer.
J.Biol.Chem., 276, 2001
|
|
1J9Z
 
 | | CYPOR-W677G | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Hubbard, P.A, Shen, A.L, Paschke, R, Kasper, C.B, Kim, J.J. | | Deposit date: | 2001-05-29 | | Release date: | 2001-08-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | NADPH-cytochrome P450 oxidoreductase. Structural basis for hydride and electron transfer.
J.Biol.Chem., 276, 2001
|
|
1JA1
 
 | | CYPOR-Triple Mutant | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Hubbard, P.A, Shen, A.L, Paschke, R, Kasper, C.B, Kim, J.J. | | Deposit date: | 2001-05-29 | | Release date: | 2001-08-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | NADPH-cytochrome P450 oxidoreductase. Structural basis for hydride and electron transfer.
J.Biol.Chem., 276, 2001
|
|
1PS9
 
 | | The Crystal Structure and Reaction Mechanism of E. coli 2,4-Dienoyl CoA Reductase | | Descriptor: | 2,4-dienoyl-CoA reductase, 5-MERCAPTOETHANOL-2-DECENOYL-COENZYME A, CHLORIDE ION, ... | | Authors: | Hubbard, P.A, Liang, X, Schulz, H, Kim, J.J. | | Deposit date: | 2003-06-20 | | Release date: | 2003-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure and reaction mechanism of Escherichia coli 2,4-dienoyl-CoA reductase
J.Biol.Chem., 278, 2003
|
|
4QT5
 
 | | Crystal Structure of 3BD10: A Monoclonal Antibody against the TSH Receptor | | Descriptor: | 3BD10 mouse monoclonal antibody, heavy chain, light chain | | Authors: | Hubbard, P.A, Chen, C.R, McLachlan, S.M, Rapoport, B, Murali, R. | | Deposit date: | 2014-07-07 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a TSH receptor monoclonal antibody: insight into Graves' disease pathogenesis.
Mol.Endocrinol., 29, 2015
|
|
3ZTN
 
 | | STRUCTURE OF INFLUENZA A NEUTRALIZING ANTIBODY SELECTED FROM CULTURES OF SINGLE HUMAN PLASMA CELLS IN COMPLEX WITH HUMAN H1 INFLUENZA HAEMAGGLUTININ. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FI6V3 ANTIBODY LIGHT CHAIN, ... | | Authors: | Hubbard, P.A, Ritchie, A.J, Corti, D, Voss, J.E, Gamblin, S.J, Codoni, G, Macagno, A, Jarrossay, D, Pinna, D, Minola, A, Vanzetta, F, Silacci, C, Fernandez-Rodriguez, B.M, Agatic, G, Giacchetto-Sasselli, I, Vachieri, S.G, Sallusto, F, Collins, P.J, Haire, L.F, Temperton, N, Langedijk, J.P.M, Skehel, J.J, Lanzavecchia, A. | | Deposit date: | 2011-07-12 | | Release date: | 2011-08-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | A Neutralizing Antibody Selected from Plasma Cells that Binds to Group 1 and Group 2 Influenza a Hemagglutinins.
Science, 333, 2011
|
|
4XC8
 
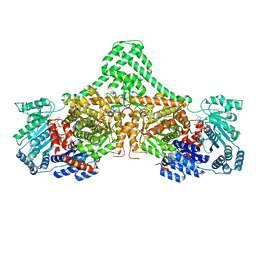 | | Isobutyryl-CoA mutase fused with bound butyryl-CoA, GDP, and Mg and without cobalamin (apo-IcmF/GDP) | | Descriptor: | Butyryl Coenzyme A, GUANOSINE-5'-DIPHOSPHATE, Isobutyryl-CoA mutase fused, ... | | Authors: | Jost, M, Drennan, C.L. | | Deposit date: | 2014-12-17 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Visualization of a radical B12 enzyme with its G-protein chaperone.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4HCY
 
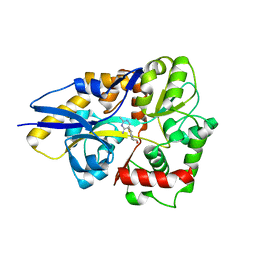 | | Structure of a eukaryotic thiaminase-I bound to the thiamin analogue 3-deazathiamin | | Descriptor: | 2-{4-[(4-amino-2-methylpyrimidin-5-yl)methyl]-3-methylthiophen-2-yl}ethanol, thiaminase-I | | Authors: | Kreinbring, C.A, Hubbard, P.A, Leeper, F.J, Hawksley, D, Petsko, G.A, Ringe, D. | | Deposit date: | 2012-10-01 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of a eukaryotic thiaminase I.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4XC7
 
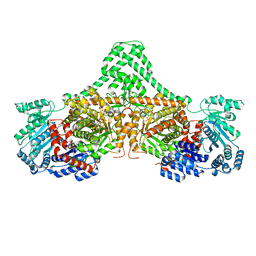 | |
4XC6
 
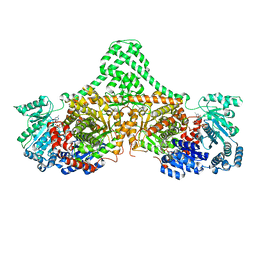 | | Isobutyryl-CoA mutase fused with bound adenosylcobalamin, GDP, and Mg (holo-IcmF/GDP) | | Descriptor: | 5'-DEOXYADENOSINE, ACETATE ION, COBALAMIN, ... | | Authors: | Jost, M, Drennan, C.L. | | Deposit date: | 2014-12-17 | | Release date: | 2015-02-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Visualization of a radical B12 enzyme with its G-protein chaperone.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4HCW
 
 | |
4H2D
 
 | | Crystal structure of NDOR1 | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADPH-dependent diflavin oxidoreductase 1 | | Authors: | Banci, L, Bertini, I, Calderone, V, Ciofi-Baffoni, S, Mikolajczyk, M, Jaiswal, D, Winkelmann, J. | | Deposit date: | 2012-09-12 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular view of an electron transfer process essential for iron-sulfur protein biogenesis.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
