5C5A
 
 | | Crystal Structure of HDM2 in complex with Nutlin-3a | | Descriptor: | 4-({(4S,5R)-4,5-bis(4-chlorophenyl)-2-[4-methoxy-2-(propan-2-yloxy)phenyl]-4,5-dihydro-1H-imidazol-1-yl}carbonyl)piperazin-2-one, CHLORIDE ION, E3 ubiquitin-protein ligase Mdm2, ... | | Authors: | Orts, J, Waelti, M.A, Marsh, M, Vera, L, Gossert, A.D, Guentert, P, Riek, R. | | Deposit date: | 2015-06-19 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.146 Å) | | Cite: | NMR Molecular Replacement, NMR2
To Be Published
|
|
7EN4
 
 | | Multi-state structure determination and dynamics analysis elucidate a new ubiquitin-recognition mechanism of yeast ubiquitin C-terminal hydrolase. | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase YUH1 | | Authors: | Okada, M, Tateishi, Y, Nojiri, E, Mikawa, T, Rajesh, S, Ogasawa, H, Ueda, T, Yagi, H, Kohno, T, Kigawa, T, Shimada, I, Guentert, P, Yutaka, I, Ikeya, T. | | Deposit date: | 2021-04-15 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Multi-state structure determination and dynamics analysis elucidate a new ubiquitin-recognition mechanism of yeast ubiquitin C-terminal hydrolase.
To Be Published
|
|
8OVI
 
 | | BRICHOS monomer | | Descriptor: | Pulmonary surfactant-associated protein C (Fragment) | | Authors: | Ghosh, D, Torres, F, Guentert, P, Riek, R. | | Deposit date: | 2023-04-26 | | Release date: | 2024-06-26 | | Method: | SOLUTION NMR | | Cite: | The inhibitory action of the chaperone BRICHOS against the alpha-Synuclein secondary nucleation pathway at near-atomic resolution
To Be Published
|
|
8OX2
 
 | | BRICHOS trimer | | Descriptor: | Pulmonary surfactant-associated protein C | | Authors: | Ghosh, D, Torres, F, Guentert, P, Riek, R. | | Deposit date: | 2023-04-30 | | Release date: | 2024-06-26 | | Method: | SOLUTION NMR | | Cite: | The inhibitory action of the chaperone BRICHOS against the alpha-Synuclein secondary nucleation pathway at near-atomic resolution
To Be Published
|
|
1MO8
 
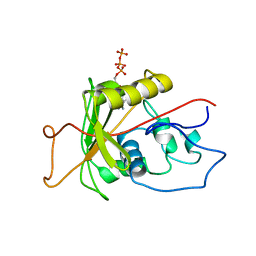 | | ATPase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Sodium/Potassium-Transporting ATPase alpha-1 | | Authors: | Hilge, M, Siegal, G, Vuister, G.W, Guentert, P, Gloor, S.M, Abrahams, J.P. | | Deposit date: | 2002-09-08 | | Release date: | 2003-06-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | ATP-induced conformational changes of the nucleotide-binding domain of Na,K-ATPase
Nat.Struct.Biol., 10, 2003
|
|
1MO7
 
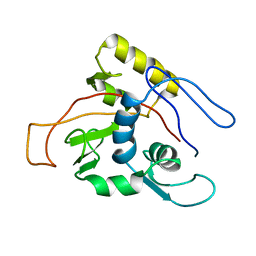 | | ATPase | | Descriptor: | Sodium/Potassium-transporting ATPase alpha-1 chain | | Authors: | Hilge, M, Siegal, G, Vuister, G.W, Guentert, P, Gloor, S.M, Abrahams, J.P. | | Deposit date: | 2002-09-08 | | Release date: | 2003-06-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | ATP-induced conformational changes of the nucleotide-binding domain of Na,K-ATPase
Nat.Struct.Biol., 10, 2003
|
|
2H3S
 
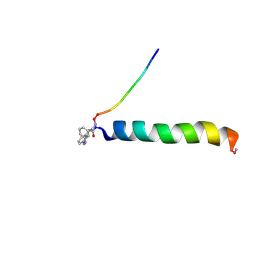 | | cis-Azobenzene-avian pancreatic polypeptide bound to DPC micelles | | Descriptor: | (3-{(Z)-[3-(AMINOMETHYL)PHENYL]DIAZENYL}PHENYL)ACETIC ACID, Pancreatic hormone | | Authors: | Jurt, S, Aemissegger, A, Guentert, P, Zerbe, O, Hilvert, D. | | Deposit date: | 2006-05-23 | | Release date: | 2006-09-12 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | A Photoswitchable Miniprotein Based on the Sequence of Avian Pancreatic Polypeptide
Angew.Chem.Int.Ed.Engl., 45, 2006
|
|
2H3T
 
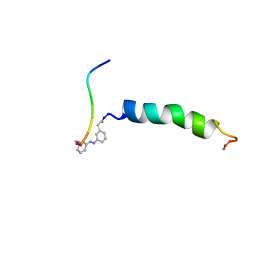 | | trans-(4-aminomethyl)phenylazobenzoic acid-aPP bound to DPC micelles | | Descriptor: | (3-{(E)-[3-(AMINOMETHYL)PHENYL]DIAZENYL}PHENYL)ACETIC ACID, Pancreatic hormone | | Authors: | Jurt, S, Aemissegger, A, Guentert, P, Zerbe, O, Hilvert, D. | | Deposit date: | 2006-05-23 | | Release date: | 2006-09-12 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | A Photoswitchable Miniprotein Based on the Sequence of Avian Pancreatic Polypeptide
Angew.Chem.Int.Ed.Engl., 45, 2006
|
|
2H4B
 
 | | cis-4-aminomethylphenylazobenzoic acid-avian pancreatic polypeptide | | Descriptor: | (3-{(Z)-[3-(AMINOMETHYL)PHENYL]DIAZENYL}PHENYL)ACETIC ACID, Pancreatic hormone | | Authors: | Jurt, S, Aemissegger, A, Guentert, P, Zerbe, O, Hilvert, D. | | Deposit date: | 2006-05-24 | | Release date: | 2006-09-12 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | A Photoswitchable Miniprotein Based on the Sequence of Avian Pancreatic Polypeptide
Angew.Chem.Int.Ed.Engl., 45, 2006
|
|
1JAS
 
 | | HsUbc2b | | Descriptor: | UBIQUITIN-CONJUGATING ENZYME E2-17 KDA | | Authors: | Miura, T, Klaus, W, Ross, A, Guentert, P, Senn, H. | | Deposit date: | 2001-05-31 | | Release date: | 2003-09-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the class I human ubiquitin-conjugating enzyme 2b
J.Biomol.NMR, 22, 2002
|
|
2DT6
 
 | | Solution structure of the first SURP domain of human splicing factor SF3a120 | | Descriptor: | Splicing factor 3 subunit 1 | | Authors: | Kuwasako, K, He, F, Inoue, M, Tanaka, A, Sugano, S, Guentert, P, Muto, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-07-11 | | Release date: | 2006-12-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the SURP domains and the subunit-assembly mechanism within the splicing factor SF3a complex in 17S U2 snRNP
Structure, 14, 2006
|
|
2N9K
 
 | | 1H, 13C, and 15N Chemical Shift Assignments for in vitro GB1 | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Ikeya, T, Hanashima, T, Hosoya, S, Shimazaki, M, Ikeda, S, Mishima, M, Guentert, P, Ito, Y. | | Deposit date: | 2015-11-26 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Improved in-cell structure determination of proteins at near-physiological concentration
Sci Rep, 6, 2016
|
|
2N9L
 
 | | 1H, 13C, and 15N Chemical Shift Assignments for in-cell GB1 | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Ikeya, T, Hanashima, T, Hosoya, S, Shimazaki, M, Ikeda, S, Mishima, M, Guentert, P, Ito, Y. | | Deposit date: | 2015-11-30 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Improved in-cell structure determination of proteins at near-physiological concentration
Sci Rep, 6, 2016
|
|
1WLN
 
 | |
4MRT
 
 | | Structure of the Phosphopantetheine Transferase Sfp in Complex with Coenzyme A and a Peptidyl Carrier Protein | | Descriptor: | 4'-phosphopantetheinyl transferase sfp, COENZYME A, GLYCEROL, ... | | Authors: | Tufar, P, Rahighi, S, Kraas, F.I, Kirchner, D.K, Loehr, F, Henrich, E, Koepke, J, Dikic, I, Guentert, P, Marahiel, M.A, Doetsch, V. | | Deposit date: | 2013-09-17 | | Release date: | 2014-04-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a PCP/Sfp Complex Reveals the Structural Basis for Carrier Protein Posttranslational Modification.
Chem.Biol., 21, 2014
|
|
1QLQ
 
 | | Bovine Pancreatic Trypsin Inhibitor (BPTI) Mutant with Altered Binding Loop Sequence | | Descriptor: | PANCREATIC TRYPSIN INHIBITOR, SULFATE ION | | Authors: | Czapinska, H, Krzywda, S, Sheldrick, G.M, Otlewski, J, Jaskolski, M. | | Deposit date: | 1999-09-10 | | Release date: | 1999-10-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | High Resolution Structure of Bovine Pancreatic Trypsin Inhibitor with Altered Binding Loop Sequence
J.Mol.Biol., 295, 1999
|
|
7QCX
 
 | | Two-state liquid NMR Structure of a PDZ2 Domain from hPTP1E, apo form | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 13 | | Authors: | Ashkinadze, D, Kadavath, H, Chi, C, Friedmann, M, Strotz, D, Kumari, P, Minges, M, Cadalbert, R, Koenigl, S, Guentert, P, Voegeli, B, Riek, R. | | Deposit date: | 2021-11-25 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Atomic resolution protein allostery from the multi-state structure of a PDZ domain.
Nat Commun, 13, 2022
|
|
7QCY
 
 | | Two-state liquid NMR Structure of a PDZ2 Domain from hPTP1E, complexed with RA-GEF2 peptide | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 13 | | Authors: | Ashkinadze, D, Kadavath, H, Chi, C, Friedmann, M, Strotz, D, Kumari, P, Minges, M, Cadalbert, R, Koenigl, S, Guentert, P, Voegeli, B, Riek, R. | | Deposit date: | 2021-11-25 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Atomic resolution protein allostery from the multi-state structure of a PDZ domain.
Nat Commun, 13, 2022
|
|
6TUB
 
 | | Beta-endorphin amyloid fibril | | Descriptor: | Beta-endorphin | | Authors: | Verasdonck, J, Seuring, C, Gath, J, Ghosh, D, Nespovitaya, N, Waelti, M.A, Maji, S, Cadalbert, R, Boeckmann, A, Guentert, P, Meier, B.H, Riek, R. | | Deposit date: | 2020-01-05 | | Release date: | 2020-10-28 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | The three-dimensional structure of human beta-endorphin amyloid fibrils.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5N2O
 
 | | Structure Of P63 SAM Domain L514F Mutant Causative Of AEC Syndrome | | Descriptor: | Tumor protein 63 | | Authors: | Rinnenthal, J, Wuerz, J.M, Osterburg, C, Guentert, P, Doetsch, V. | | Deposit date: | 2017-02-08 | | Release date: | 2018-02-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Protein aggregation of the p63 transcription factor underlies severe skin fragility in AEC syndrome.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6H8C
 
 | | Structure of the human GABARAPL2 protein in complex with the UBA5 LIR motif | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein-like 2, Ubiquitin-like modifier-activating enzyme 5 | | Authors: | Huber, J, Loehr, F, Gruber, J, Akutsu, M, Guentert, P, Doetsch, V, Rogov, V.V. | | Deposit date: | 2018-08-02 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | An atypical LIR motif within UBA5 (ubiquitin like modifier activating enzyme 5) interacts with GABARAP proteins and mediates membrane localization of UBA5.
Autophagy, 16, 2020
|
|
5ZCZ
 
 | | Solution structure of the T. Thermophilus HB8 TTHA1718 protein in living eukaryotic cells by in-cell NMR spectroscopy | | Descriptor: | Heavy metal binding protein | | Authors: | Tanaka, T, Teppei, I, Kamoshida, H, Mishima, M, Shirakawa, M, Guentert, P, Ito, Y. | | Deposit date: | 2018-02-22 | | Release date: | 2019-08-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Protein 3D Structure Determination in Living Eukaryotic Cells.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
5ZD0
 
 | | Solution structure of human ubiquitin with three alanine mutations in living eukaryotic cells by in-cell NMR spectroscopy | | Descriptor: | ubiquitin | | Authors: | Tanaka, T, Ikeya, T, Kamoshida, H, Mishima, M, Shirakawa, M, Guentert, P, Ito, Y. | | Deposit date: | 2018-02-22 | | Release date: | 2019-08-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Protein 3D Structure Determination in Living Eukaryotic Cells.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
5Z4B
 
 | | GB1 structure determination in living eukaryotic cells by in-cell NMR spectroscopy | | Descriptor: | Protein LG | | Authors: | Tanaka, T, Teppei, I, Kamoshida, H, Mishima, M, Shirakawa, M, Guentert, P, Ito, Y. | | Deposit date: | 2018-01-10 | | Release date: | 2019-01-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Protein 3D Structure Determination in Living Eukaryotic Cells.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
2RON
 
 | | The external thioesterase of the Surfactin-Synthetase | | Descriptor: | Surfactin synthetase thioesterase subunit | | Authors: | Koglin, A, Lohr, F, Bernhard, F, Rogov, V.V, Frueh, D.P, Strieter, E.R, Mofid, M.R, Guentert, P, Wagner, G, Walsh, C.T, Marahiel, M.A, Doetsch, V. | | Deposit date: | 2008-04-04 | | Release date: | 2008-08-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the selectivity of the external thioesterase of the surfactin synthetase
Nature, 454, 2008
|
|
