158D
 
 | | CRYSTALLOGRAPHIC ANALYSIS OF C-C-A-A-G-C-T-T-G-G AND ITS IMPLICATIONS FOR BENDING IN B-DNA | | Descriptor: | CALCIUM ION, DNA (5'-D(*CP*CP*AP*AP*GP*CP*TP*TP*GP*G)-3') | | Authors: | Grzeskowiak, K, Goodsell, D.S, Kaczor-Grzeskowiak, M, Cascio, D, Dickerson, R.E. | | Deposit date: | 1994-02-03 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic analysis of C-C-A-A-G-C-T-T-G-G and its implications for bending in B-DNA.
Biochemistry, 32, 1993
|
|
1D23
 
 | | THE STRUCTURE OF B-HELICAL C-G-A-T-C-G-A-T-C-G AND COMPARISON WITH C-C-A-A-C-G-T-T-G-G. THE EFFECT OF BASE PAIR REVERSALS | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*CP*GP*AP*TP*CP*G)-3'), MAGNESIUM ION | | Authors: | Grzeskowiak, K, Yanagi, K, Prive, G.G, Dickerson, R.E. | | Deposit date: | 1991-05-29 | | Release date: | 1991-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of B-helical C-G-A-T-C-G-A-T-C-G and comparison with C-C-A-A-C-G-T-T-G-G. The effect of base pair reversals.
J.Biol.Chem., 266, 1991
|
|
1RNL
 
 | | THE NITRATE/NITRITE RESPONSE REGULATOR PROTEIN NARL FROM NARL | | Descriptor: | GLYCEROL, NITRATE/NITRITE RESPONSE REGULATOR PROTEIN NARL, PLATINUM (II) ION | | Authors: | Baikalov, I, Schroder, I, Kaczor-Grzeskowiak, M, Grzeskowiak, K, Gunsalus, R.P, Dickerson, R.E. | | Deposit date: | 1996-04-17 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Escherichia coli response regulator NarL.
Biochemistry, 35, 1996
|
|
2IE1
 
 | | Polyamines stabilize left-handed Z-DNA. We found new type of polyamine which stabilize left-handed Z-DNA by X-ray crystallography | | Descriptor: | DNA (5'-D(*DCP*DGP*DCP*DGP*DCP*DG)-3'), N-(2-AMINOETHYL)-N'-{2-[(2-AMINOETHYL)AMINO]ETHYL}ETHANE-1,2-DIAMINE | | Authors: | Ohishi, H, Odoko, M, Tsukamoto, K, Hiyama, Y, Maezaki, N, Grzeskowiak, K, Ishida, T, Tanaka, T, Okabe, N, Fukuyama, K. | | Deposit date: | 2006-09-16 | | Release date: | 2007-10-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Polyamines stabilize left-handed Z-DNA. We found new type of polyamine which stabilize left-handed Z-DNA by X-ray crystallography
To be Published
|
|
8BNA
 
 | | BINDING OF HOECHST 33258 TO THE MINOR GROOVE OF B-DNA | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Pjura, P.E, Grzeskowiak, K, Dickerson, R.E. | | Deposit date: | 1986-08-29 | | Release date: | 1987-01-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Binding of Hoechst 33258 to the minor groove of B-DNA.
J.Mol.Biol., 197, 1987
|
|
307D
 
 | | Structure of a DNA analog of the primer for HIV-1 RT second strand synthesis | | Descriptor: | DNA (5'-D(*CP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3'), DNA (5'-D(*CP*TP*TP*TP*TP*CP*TP*TP*TP*G)-3') | | Authors: | Han, G.W, Kopka, M.L, Cascio, D, Grzeskowiak, K, Dickerson, R.E. | | Deposit date: | 1997-01-07 | | Release date: | 1997-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of a DNA analog of the primer for HIV-1 RT second strand synthesis.
J.Mol.Biol., 269, 1997
|
|
196D
 
 | |
274D
 
 | | CRYSTAL STRUCTURE OF A COVALENT DNA-DRUG ADDUCT: ANTHRAMYCIN BOUND TO C-C-A-A-C-G-T-T-G-G, AND A MOLECULAR EXPLANATION OF SPECIFICITY | | Descriptor: | ANTHRAMYCIN, DNA (5'-D(*CP*CP*AP*AP*CP*GP*TP*TP*(DRUG)GP*G)-3') | | Authors: | Kopka, M.L, Goodsell, D.S, Baikalov, I, Grzeskowiak, K, Cascio, D, Dickerson, R.E. | | Deposit date: | 1994-04-28 | | Release date: | 1994-10-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a covalent DNA-drug adduct: anthramycin bound to C-C-A-A-C-G-T-T-G-G and a molecular explanation of specificity.
Biochemistry, 33, 1994
|
|
1D49
 
 | | THE STRUCTURE OF A B-DNA DECAMER WITH A CENTRAL T-A STEP: C-G-A-T-T-A-A-T-C-G | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*TP*AP*AP*TP*CP*G)-3'), MAGNESIUM ION | | Authors: | Quintana, J.R, Grzeskowiak, K, Yanagi, K, Dickerson, R.E. | | Deposit date: | 1991-09-17 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a B-DNA decamer with a central T-A step: C-G-A-T-T-A-A-T-C-G.
J.Mol.Biol., 225, 1992
|
|
1DA3
 
 | | THE CRYSTAL STRUCTURE OF THE TRIGONAL DECAMER C-G-A-T-C-G-6MEA-T-C-G: A B-DNA HELIX WITH 10.6 BASE-PAIRS PER TURN | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*GP*AP*TP*CP*GP*(6MA)P*TP*CP*G)-3'), MAGNESIUM ION | | Authors: | Baikalov, I, Grzeskowiak, K, Yanagi, K, Quintana, J, Dickerson, R.E. | | Deposit date: | 1992-11-09 | | Release date: | 1993-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the trigonal decamer C-G-A-T-C-G-6meA-T-C-G: a B-DNA helix with 10.6 base-pairs per turn.
J.Mol.Biol., 231, 1993
|
|
1D16
 
 | |
1D30
 
 | | THE STRUCTURE OF DAPI BOUND TO DNA | | Descriptor: | 6-AMIDINE-2-(4-AMIDINO-PHENYL)INDOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Larsen, T, Goodsell, D.S, Cascio, D, Grzeskowiak, K, Dickerson, R.E. | | Deposit date: | 1991-01-04 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of DAPI bound to DNA.
J.Biomol.Struct.Dyn., 7, 1989
|
|
1A04
 
 | | THE STRUCTURE OF THE NITRATE/NITRITE RESPONSE REGULATOR PROTEIN NARL IN THE MONOCLINIC C2 CRYSTAL FORM | | Descriptor: | NITRATE/NITRITE RESPONSE REGULATOR PROTEIN NARL | | Authors: | Baikalov, I, Schroder, I, Kaczor-Grzeskowiak, M, Cascio, D, Gunsalus, R.P, Dickerson, R.E. | | Deposit date: | 1997-12-08 | | Release date: | 1998-03-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | NarL dimerization? Suggestive evidence from a new crystal form
Biochemistry, 37, 1998
|
|
1ZG5
 
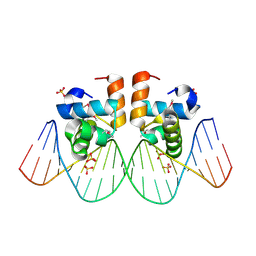 | | NarL complexed to narG-89 promoter palindromic tail-to-tail DNA site | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*CP*CP*CP*TP*AP*TP*AP*GP*GP*GP*GP*TP*AP*CP*G)-3', Nitrate/nitrite response regulator protein narL, SULFATE ION | | Authors: | Maris, A.E, Kaczor-Grzeskowiak, M, Ma, Z, Kopka, M.L, Gunsalus, R.P, Dickerson, R.E. | | Deposit date: | 2005-04-20 | | Release date: | 2005-11-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Primary and Secondary Modes of DNA Recognition by the NarL Two-Component Response Regulator.
Biochemistry, 44, 2005
|
|
1ZG1
 
 | | NarL complexed to nirB promoter non-palindromic tail-to-tail DNA site | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*CP*CP*AP*TP*TP*AP*AP*GP*GP*AP*GP*TP*AP*CP*G)-3', 5'-D(*CP*GP*TP*AP*CP*TP*CP*CP*TP*TP*AP*AP*TP*GP*GP*GP*TP*AP*CP*G)-3', Nitrate/nitrite response regulator protein narL, ... | | Authors: | Maris, A.E, Kaczor-Grzeskowiak, M, Ma, Z, Kopka, M.L, Gunsalus, R.P, Dickerson, R.E. | | Deposit date: | 2005-04-20 | | Release date: | 2005-11-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Primary and Secondary Modes of DNA Recognition by the NarL Two-Component Response Regulator.
Biochemistry, 44, 2005
|
|
1SK5
 
 | |
1D56
 
 | |
1D57
 
 | |
252D
 
 | |
1D43
 
 | | DNA DODECAMER C-G-C-G-A-A-T-T-C-G-C-G/HOECHST 33258 COMPLEX: 0 DEGREES C, PIPERAZINE UP | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Quintana, J.R, Lipanov, A.A, Dickerson, R.E. | | Deposit date: | 1991-06-04 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Low-temperature crystallographic analyses of the binding of Hoechst 33258 to the double-helical DNA dodecamer C-G-C-G-A-A-T-T-C-G-C-G.
Biochemistry, 30, 1991
|
|
1D46
 
 | | DNA DODECAMER C-G-C-G-A-A-T-T-C-G-C-G/HOECHST 33258 COMPLEX:-100 DEGREES C, PIPERAZINE DOWN | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Quintana, J.R, Lipanov, A.A, Dickerson, R.E. | | Deposit date: | 1991-06-04 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Low-temperature crystallographic analyses of the binding of Hoechst 33258 to the double-helical DNA dodecamer C-G-C-G-A-A-T-T-C-G-C-G.
Biochemistry, 30, 1991
|
|
1D45
 
 | | DNA DODECAMER C-G-C-G-A-A-T-T-C-G-C-G/HOECHST 33258 COMPLEX:-25 DEGREES C, PIPERAZINE DOWN | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Quintana, J.R, Lipanov, A.A, Dickerson, R.E. | | Deposit date: | 1991-06-04 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Low-temperature crystallographic analyses of the binding of Hoechst 33258 to the double-helical DNA dodecamer C-G-C-G-A-A-T-T-C-G-C-G.
Biochemistry, 30, 1991
|
|
1D44
 
 | | DNA DODECAMER C-G-C-G-A-A-T-T-C-G-C-G/HOECHST 33258 COMPLEX: 0 DEGREES C, PIPERAZINE DOWN | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Quintana, J.R, Lipanov, A.A, Dickerson, R.E. | | Deposit date: | 1991-06-04 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Low-temperature crystallographic analyses of the binding of Hoechst 33258 to the double-helical DNA dodecamer C-G-C-G-A-A-T-T-C-G-C-G.
Biochemistry, 30, 1991
|
|
