3MMH
 
 | | X-ray structure of free methionine-R-sulfoxide reductase from neisseria meningitidis in complex with its substrate | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Gruez, A, Libiad, M, Boschi-Muller, S, Branlant, G. | | Deposit date: | 2010-04-19 | | Release date: | 2010-05-19 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural and Biochemical Characterization of Free Methionine-R-sulfoxide Reductase from Neisseria meningitidis.
J.Biol.Chem., 285, 2010
|
|
1DDI
 
 | | CRYSTAL STRUCTURE OF SIR-FP60 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFITE REDUCTASE [NADPH] FLAVOPROTEIN ALPHA-COMPONENT | | Authors: | Gruez, A, Pignol, D, Zeghouf, M, Coves, J, Fontecave, M, Ferrer, J.L, Fontecilla-Camps, J.C. | | Deposit date: | 1999-11-10 | | Release date: | 2000-11-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Four crystal structures of the 60 kDa flavoprotein monomer of the sulfite reductase indicate a disordered flavodoxin-like module.
J.Mol.Biol., 299, 2000
|
|
1DDG
 
 | | CRYSTAL STRUCTURE OF SIR-FP60 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, SULFITE REDUCTASE (NADPH) FLAVOPROTEIN ALPHA-COMPONENT | | Authors: | Gruez, A, Pignol, D, Zeghouf, M, Coves, J, Fontecave, M, Ferrer, J.L, Fontecilla-Camps, J.C. | | Deposit date: | 1999-11-10 | | Release date: | 2000-11-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Four crystal structures of the 60 kDa flavoprotein monomer of the sulfite reductase indicate a disordered flavodoxin-like module.
J.Mol.Biol., 299, 2000
|
|
1WND
 
 | | Escherichia coli YdcW gene product is a medium-chain aldehyde dehydrogenase as determined by kinetics and crystal structure | | Descriptor: | CALCIUM ION, Putative betaine aldehyde dehydrogenase | | Authors: | Gruez, A, Roig-Zamboni, V, Tegoni, M, Cambillau, C. | | Deposit date: | 2004-07-29 | | Release date: | 2004-10-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure and Kinetics Identify Escherichia coli YdcW Gene Product as a Medium-chain Aldehyde Dehydrogenase
J.Mol.Biol., 343, 2004
|
|
1WNB
 
 | | Escherichia coli YdcW gene product is a medium-chain aldehyde dehydrogenase (complexed with nadh and betaine aldehyde) | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, BETAINE ALDEHYDE, Putative betaine aldehyde dehydrogenase | | Authors: | Gruez, A, Roig-Zamboni, V, Tegoni, M, Cambillau, C. | | Deposit date: | 2004-07-29 | | Release date: | 2004-10-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure and Kinetics Identify Escherichia coli YdcW Gene Product as a Medium-chain Aldehyde Dehydrogenase
J.Mol.Biol., 343, 2004
|
|
3CDU
 
 | | Crystal structure of coxsackievirus B3 RNA-dependent RNA polymerase (3Dpol) in complex with a pyrophosphate | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Gruez, A, Selisko, B, Roberts, M, Bricogne, G, Bussetta, C, Canard, B. | | Deposit date: | 2008-02-27 | | Release date: | 2008-07-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of coxsackievirus B3 RNA-dependent RNA polymerase in complex with its protein primer VPg confirms the existence of a second VPg binding site on Picornaviridae polymerases
J.Virol., 82, 2008
|
|
3CDW
 
 | | Crystal structure of coxsackievirus B3 RNA-dependent RNA polymerase (3Dpol) in complex with protein primer VPg and a pyrophosphate | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Gruez, A, Selisko, B, Roberts, M, Bricogne, G, Bussetta, C, Canard, B. | | Deposit date: | 2008-02-27 | | Release date: | 2008-07-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of coxsackievirus B3 RNA-dependent RNA polymerase in complex with its protein primer VPg confirms the existence of a second VPg binding site on Picornaviridae polymerases
J.Virol., 82, 2008
|
|
1PT7
 
 | | Crystal structure of the apo-form of the yfdW gene product of E. coli | | Descriptor: | GLYCEROL, Hypothetical protein yfdW, PHOSPHATE ION | | Authors: | Gruez, A, Roig-Zamboni, V, Valencia, C, Campanacci, V, Cambillau, C. | | Deposit date: | 2003-06-23 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of the Escherichia coli yfdW gene product reveals a New fold of two interlaced rings identifying a wide family of CoA transferases.
J.Biol.Chem., 278, 2003
|
|
1PT8
 
 | | Crystal structure of the yfdW gene product of E. coli, in complex with oxalate and acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, GLYCEROL, Hypothetical protein yfdW, ... | | Authors: | Gruez, A, Roig-Zamboni, V, Valencia, C, Campanacci, V, Cambillau, C. | | Deposit date: | 2003-06-23 | | Release date: | 2003-09-09 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of the Escherichia coli yfdW gene product reveals a new fold of two interlaced rings identifying a wide family of CoA transferases.
J.Biol.Chem., 278, 2003
|
|
1PT5
 
 | | Crystal structure of gene yfdW of E. coli | | Descriptor: | ACETYL COENZYME *A, Hypothetical protein yfdW | | Authors: | Gruez, A, Roig-Zamboni, V, Valencia, C, Campanacci, V, Cambillau, C. | | Deposit date: | 2003-06-23 | | Release date: | 2003-09-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of the Escherichia coli YfdW Gene Product Reveals a New Fold of Two Interlaced Rings Identifying a Wide Family of CoA Transferases
J.Biol.Chem., 278, 2003
|
|
6F7L
 
 | | Crystal structure of LkcE R326Q mutant in complex with its substrate | | Descriptor: | ACETATE ION, Amine oxidase LkcE, CALCIUM ION, ... | | Authors: | Dorival, J, Risser, F, Jacob, C, Collin, S, Drager, G, Kirschning, A, Paris, C, Chagot, B, Gruez, A, Weissman, K.J. | | Deposit date: | 2017-12-11 | | Release date: | 2018-09-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into a dual function amide oxidase/macrocyclase from lankacidin biosynthesis.
Nat Commun, 9, 2018
|
|
2VJQ
 
 | | Formyl-CoA transferase mutant variant W48Q | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FORMYL-COENZYME A TRANSFERASE | | Authors: | Toyota, C.G, Berthold, C.L, Gruez, A, Jonsson, S, Lindqvist, Y, Cambillau, C, Richards, N.G.J. | | Deposit date: | 2007-12-11 | | Release date: | 2008-01-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Differential Substrate Specificity and Kinetic Behavior of Escherichia Coli Yfdw and Oxalobacter Formigenes Formyl Coenzyme a Transferase.
J.Bacteriol., 190, 2008
|
|
8ODW
 
 | | Crystal structure of LbmA Ox-ACP didomain in complex with NADP and ethyl glycinate from the lobatamide PKS (Gynuella sunshinyii) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Francois, R.M.M, Fraley, A.E, Piel, J, Weissman, K.J, Gruez, A. | | Deposit date: | 2023-03-09 | | Release date: | 2023-05-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Modular Oxime Formation by a trans-AT Polyketide Synthase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8QZU
 
 | |
2VJP
 
 | | Formyl-CoA transferase mutant variant W48F | | Descriptor: | FORMYL-COENZYME A TRANSFERASE, SODIUM ION | | Authors: | Toyota, C.G, Berthold, C.L, Gruez, A, Jonsson, S, Lindqvist, Y, Cambillau, C, Richards, N.G.J. | | Deposit date: | 2007-12-11 | | Release date: | 2008-01-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Differential Substrate Specificity and Kinetic Behavior of Escherichia Coli Yfdw and Oxalobacter Formigenes Formyl Coenzyme a Transferase.
J.Bacteriol., 190, 2008
|
|
4CA3
 
 | | SOLUTION STRUCTURE OF STREPTOMYCES VIRGINIAE VIRA ACP5B | | Descriptor: | HYBRID POLYKETIDE SYNTHASE-NON RIBOSOMAL PEPTIDE SYNTHETASE | | Authors: | Davison, J, Dorival, J, Rabeharindranto, M.H, Chagot, B, Gruez, A, Weissman, K.J. | | Deposit date: | 2013-10-05 | | Release date: | 2014-06-04 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Insights Into the Function of Trans-Acyl Transferase Polyketide Synthases from the Saxs Structure of a Complete Module.
Chem.Sci., 2014
|
|
8S6L
 
 | | AzeD dehydratase of Pseudomonas aeruginosa | | Descriptor: | GLYCEROL, Phthiocerol/phthiodiolone dimycocerosyl transferase | | Authors: | Calderari, A, Gruez, A, Najah, S, Li, Y, Weissman, K.J. | | Deposit date: | 2024-02-27 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | High resolution structure of azetidomonamide dehydratase
To be published
|
|
3BJH
 
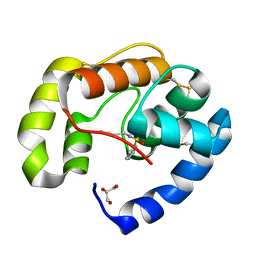 | | Soft-SAD crystal structure of a pheromone binding protein from the honeybee Apis mellifera L. | | Descriptor: | GLYCEROL, N-BUTYL-BENZENESULFONAMIDE, Pheromone-binding protein ASP1 | | Authors: | Lartigue, A, Gruez, A, Briand, L, Blon, F, Bezirard, V, Walsh, M, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2007-12-04 | | Release date: | 2007-12-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Sulfur single-wavelength anomalous diffraction crystal structure of a pheromone-binding protein from the honeybee Apis mellifera L.
J.Biol.Chem., 279, 2004
|
|
1W8G
 
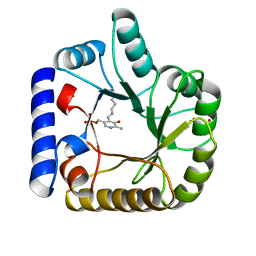 | | CRYSTAL STRUCTURE OF E. COLI K-12 YGGS | | Descriptor: | HYPOTHETICAL UPF0001 PROTEIN YGGS, ISOCITRIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Sulzenbacher, G, Gruez, A, Spinelli, S, Roig-Zamboni, V, Pagot, F, Bignon, C, Vincentelli, R, Cambillau, C. | | Deposit date: | 2004-09-21 | | Release date: | 2006-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of E. Coli K-12 Yggs
To be Published
|
|
6FJH
 
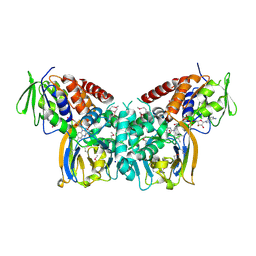 | | Crystal structure of the seleniated LkcE from Streptomyces rochei | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LkcE, OXYGEN MOLECULE, ... | | Authors: | Dorival, J, Risser, F, Jacob, C, Collin, S, Drager, G, Kirschning, A, Paris, C, Chagot, B, Gruez, A, Weissman, K.J. | | Deposit date: | 2018-01-22 | | Release date: | 2018-09-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Insights into a dual function amide oxidase/macrocyclase from lankacidin biosynthesis.
Nat Commun, 9, 2018
|
|
4LRT
 
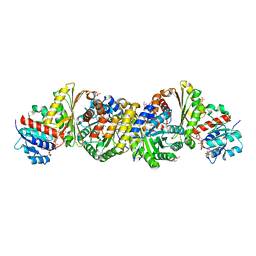 | | Crystal and solution structures of the bifunctional enzyme (Aldolase/Aldehyde dehydrogenase) from Thermomonospora curvata, reveal a cofactor-binding domain motion during NAD+ and CoA accommodation whithin the shared cofactor-binding site | | Descriptor: | 4-hydroxy-2-oxovalerate aldolase, Acetaldehyde dehydrogenase, COENZYME A, ... | | Authors: | Fischer, B, Branlant, G, Talfournier, F, Gruez, A. | | Deposit date: | 2013-07-20 | | Release date: | 2013-09-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal and solution structures of the bifunctional enzyme (Aldolase/Aldehyde dehydrogenase) from Thermomonospora curvata, reveal a cofactor-binding domain motion during NAD+ and CoA accommodation whithin the shared cofactor-binding site
To be Published
|
|
6F7V
 
 | | Crystal structure of LkcE E64Q mutant in complex with LC-KA05 | | Descriptor: | CALCIUM ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Dorival, J, Risser, F, Jacob, C, Collin, S, Drager, G, Kirschning, A, Paris, C, Chagot, B, Gruez, A, Weissman, K.J. | | Deposit date: | 2017-12-12 | | Release date: | 2018-09-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Insights into a dual function amide oxidase/macrocyclase from lankacidin biosynthesis.
Nat Commun, 9, 2018
|
|
6F32
 
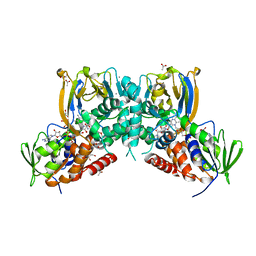 | | Crystal structure of a dual function amine oxidase/cyclase in complex with substrate analogues | | Descriptor: | (2~{R},3~{R})-2,3-bis(oxidanyl)-~{N},~{N}'-dipropyl-butanediamide, ACETATE ION, Amine oxidase LkcE, ... | | Authors: | Dorival, J, Risser, F, Jacob, C, Collin, S, Drager, G, Kirschning, A, Paris, C, Chagot, B, Gruez, A, Weissman, K.J. | | Deposit date: | 2017-11-27 | | Release date: | 2018-09-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Insights into a dual function amide oxidase/macrocyclase from lankacidin biosynthesis.
Nat Commun, 9, 2018
|
|
8S81
 
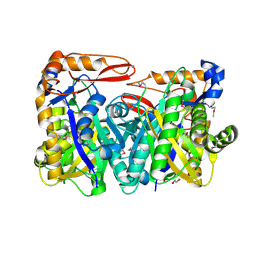 | | VirC mutant C114A-Q334A-R335A-R338A | | Descriptor: | 1,2-ETHANEDIOL, HMG-CoA synthase-like protein | | Authors: | Collin, S, Gruez, A, Weissman, K.J. | | Deposit date: | 2024-03-05 | | Release date: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Plasticity within 3-Hydroxy-3-Methylglutaryl Synthases Catalyzing the First Step of beta-Branching in Polyketide Biosynthesis Underpins a Dynamic Mechanism of Substrate Accommodation.
Jacs Au, 4, 2024
|
|
4LRS
 
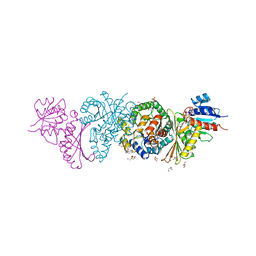 | | Crystal and solution structures of the bifunctional enzyme (Aldolase/Aldehyde dehydrogenase) from Thermomonospora curvata, reveal a cofactor-binding domain motion during NAD+ and CoA accommodation whithin the shared cofactor-binding site | | Descriptor: | 4-hydroxy-2-oxovalerate aldolase, Acetaldehyde dehydrogenase, CHLORIDE ION, ... | | Authors: | Fischer, B, Branlant, G, Talfournier, F, Gruez, A. | | Deposit date: | 2013-07-20 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal and solution structures of the bifunctional enzyme (Aldolase/Aldehyde dehydrogenase) from Thermomonospora curvata, reveal a cofactor-binding domain motion during NAD+ and CoA accommodation whithin the shared cofactor-binding site
To be Published
|
|
