1SOR
 
 | | Aquaporin-0 membrane junctions reveal the structure of a closed water pore | | Descriptor: | Aquaporin-0 | | Authors: | Gonen, T, Sliz, P, Kistler, J, Cheng, Y, Walz, T. | | Deposit date: | 2004-03-15 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | ELECTRON CRYSTALLOGRAPHY (3 Å) | | Cite: | Aquaporin-0 membrane junctions reveal the structure of a closed water pore
Nature, 429, 2004
|
|
2B6O
 
 | | Electron crystallographic structure of lens Aquaporin-0 (AQP0) (lens MIP) at 1.9A resolution, in a closed pore state | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Lens fiber major intrinsic protein | | Authors: | Gonen, T, Cheng, Y, Sliz, P, Hiroaki, Y, Fujiyoshi, Y, Harrison, S.C, Walz, T. | | Deposit date: | 2005-10-03 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.9 Å) | | Cite: | Lipid-protein interactions in double-layered two-dimensional AQP0 crystals.
Nature, 438, 2005
|
|
2B6P
 
 | | X-ray structure of lens Aquaporin-0 (AQP0) (lens MIP) in an open pore state | | Descriptor: | Lens fiber major intrinsic protein | | Authors: | Gonen, T, Cheng, Y, Sliz, P, Hiroaki, Y, Fujiyoshi, Y, Harrison, S.C, Walz, T. | | Deposit date: | 2005-10-03 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Lipid-protein interactions in double-layered two-dimensional AQP0 crystals.
Nature, 438, 2005
|
|
7RM5
 
 | | MicroED structure of the human adenosine receptor at 2.8A | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a/Soluble cytochrome b562 chimera, CHOLESTEROL, ... | | Authors: | Martynowycz, M.W, Shiriaeva, A, Ge, X, Hattne, J, Nannenga, B.L, Cherezov, V, Gonen, T. | | Deposit date: | 2021-07-26 | | Release date: | 2021-09-08 | | Last modified: | 2024-11-20 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.79 Å) | | Cite: | MicroED structure of the human adenosine receptor determined from a single nanocrystal in LCP.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6EEX
 
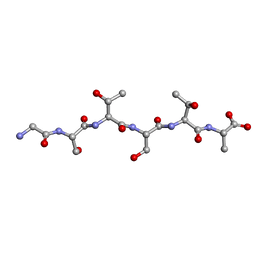 | | L-GSTSTA from degenerate octameric repeats in InaZ, residues 707-712 | | Descriptor: | L-GSTSTA from ice nucleaction protein, inaZ | | Authors: | Zee, C, Glynn, C, Gallagher-Jones, M, Miao, J, Santiago, C.G, Cascio, D, Gonen, T, Sawaya, M.R, Rodriguez, J.A. | | Deposit date: | 2018-08-15 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Homochiral and racemic MicroED structures of a peptide repeat from the ice-nucleation protein InaZ.
IUCrJ, 6, 2019
|
|
3J4R
 
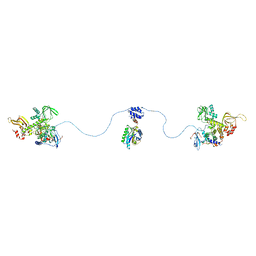 | |
3J6K
 
 | |
9ODW
 
 | |
9ODV
 
 | |
5K7P
 
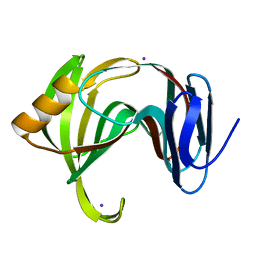 | | MicroED structure of xylanase at 2.3 A resolution | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | de la Cruz, M.J, Hattne, J, Shi, D, Seidler, P, Rodriguez, J, Reyes, F.E, Sawaya, M.R, Cascio, D, Eisenberg, D, Gonen, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-05 | | Last modified: | 2024-02-28 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.3 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
4UOT
 
 | | Thermodynamic hyperstability in parametrically designed helical bundles | | Descriptor: | DESIGNED HELICAL BUNDLE 5H2L | | Authors: | Oberdorfer, G, Huang, P, Pei, X.Y, Xu, C, Gonen, T, Nannenga, B, DiMaio, D, Rogers, J, Luisi, B.F, Baker, D. | | Deposit date: | 2014-06-09 | | Release date: | 2014-11-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | High Thermodynamic Stability of Parametrically Designed Helical Bundles
Science, 346, 2014
|
|
4UOS
 
 | | Thermodynamic hyperstability in parametrically designed helical bundles | | Descriptor: | DESIGNED HELICAL BUNDLE | | Authors: | Oberdorfer, G, Huang, P, Pei, X.Y, Xu, C, Gonen, T, Nannenga, B, DiMaio, D, Rogers, J, Luisi, B.F, Baker, D. | | Deposit date: | 2014-06-09 | | Release date: | 2014-11-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | High Thermodynamic Stability of Parametrically Designed Helical Bundles
Science, 346, 2014
|
|
8SDK
 
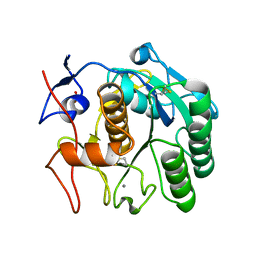 | | The MicroED structure of proteinase K crystallized by suspended drop crystallization | | Descriptor: | CALCIUM ION, Proteinase K, SULFATE ION | | Authors: | Gillman, C, Nicolas, W.J, Martynowycz, M.W, Gonen, T. | | Deposit date: | 2023-04-06 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.1 Å) | | Cite: | Design and implementation of suspended drop crystallization.
Iucrj, 10, 2023
|
|
7JSY
 
 | |
7UTE
 
 | |
7ULY
 
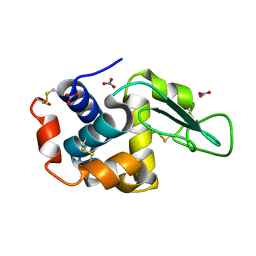 | | MicroED structure of triclinic lysozyme | | Descriptor: | Lysozyme C, NITRATE ION | | Authors: | Clabbers, M.T.B, Martynowycz, M.W, Hattne, J, Gonen, T. | | Deposit date: | 2022-04-05 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-09 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.87 Å) | | Cite: | Hydrogens and hydrogen-bond networks in macromolecular MicroED data.
J Struct Biol X, 6, 2022
|
|
9CBV
 
 | | MicroED structure of the human MP20 protein | | Descriptor: | Lens fiber membrane intrinsic protein | | Authors: | Nicolas, W.J, Shiriaeva, A, Martynowycz, M.W, Grey, A.C, Ruma, Y, Donaldson, P.J, Gonen, T. | | Deposit date: | 2024-06-20 | | Release date: | 2025-05-21 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.5 Å) | | Cite: | Structure of the lens MP20 mediated adhesive junction.
Nat Commun, 16, 2025
|
|
4JRE
 
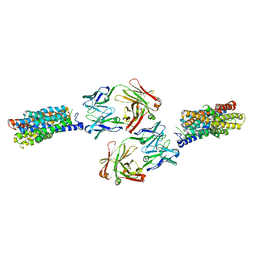 | | Crystal structure of nitrate/nitrite exchanger NarK with nitrite bound | | Descriptor: | Immunoglobulin Gamma-2a, Heavy chain, Immunoglobulin Kappa, ... | | Authors: | Zheng, H, Wisedchaisri, G, Gonen, T. | | Deposit date: | 2013-03-21 | | Release date: | 2013-05-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a nitrate/nitrite exchanger.
Nature, 497, 2013
|
|
4JR9
 
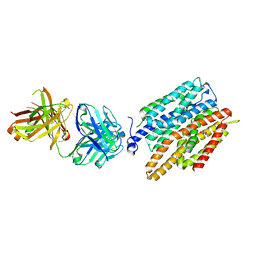 | | Crystal structure of nitrate/nitrite exchanger NarK | | Descriptor: | Immunoglobulin Gamma-2a, Heavy chain, Immunoglobulin Kappa, ... | | Authors: | Zheng, H, Wisedchaisri, G, Gonen, T. | | Deposit date: | 2013-03-21 | | Release date: | 2013-05-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a nitrate/nitrite exchanger.
Nature, 497, 2013
|
|
8EUM
 
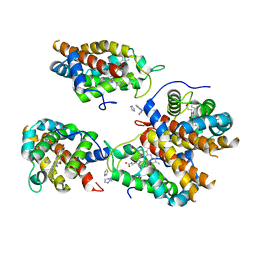 | | MicroED structure of an Aeropyrum pernix protoglobin mutant | | Descriptor: | FE (III) ION, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Danelius, E, Gonen, T, Unge, J.T. | | Deposit date: | 2022-10-18 | | Release date: | 2023-11-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.1 Å) | | Cite: | MicroED Structure of a Protoglobin Reactive Carbene Intermediate.
J.Am.Chem.Soc., 145, 2023
|
|
8E53
 
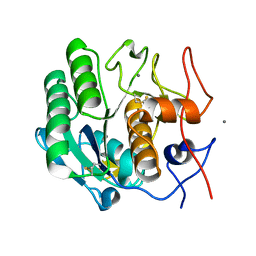 | | MicroED structure of proteinase K recorded on K3 | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Clabbers, M.T.B, Martynowycz, M.W, Hattne, J, Nannenga, B.L, Gonen, T. | | Deposit date: | 2022-08-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-11-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.7 Å) | | Cite: | Electron-counting MicroED data with the K2 and K3 direct electron detectors.
J.Struct.Biol., 214, 2022
|
|
8E52
 
 | | MicroED structure of proteinase K recorded on K2 | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Clabbers, M.T.B, Martynowycz, M.W, Hattne, J, Nannenga, B.L, Gonen, T. | | Deposit date: | 2022-08-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-11-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.8 Å) | | Cite: | Electron-counting MicroED data with the K2 and K3 direct electron detectors.
J.Struct.Biol., 214, 2022
|
|
8E54
 
 | | MicroED structure of triclinic lysozyme recorded on K3 | | Descriptor: | Lysozyme C, NITRATE ION | | Authors: | Clabbers, M.T.B, Martynowycz, M.W, Hattne, J, Nannenga, B.L, Gonen, T. | | Deposit date: | 2022-08-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.2 Å) | | Cite: | Electron-counting MicroED data with the K2 and K3 direct electron detectors.
J.Struct.Biol., 214, 2022
|
|
8EUN
 
 | | MicroED structure of an Aeropyrum pernix protoglobin metallo-carbene complex | | Descriptor: | Protogloblin ApPgb, benzyl[3,3'-(7,12-diethenyl-3,8,13,17-tetramethylporphyrin-2,18-diyl-kappa~4~N~21~,N~22~,N~23~,N~24~)di(propanoato)(2-)]iron | | Authors: | Danelius, E, Gonen, T, Unge, J.T. | | Deposit date: | 2022-10-19 | | Release date: | 2023-04-05 | | Last modified: | 2024-05-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.5 Å) | | Cite: | MicroED Structure of a Protoglobin Reactive Carbene Intermediate.
J.Am.Chem.Soc., 145, 2023
|
|
7KUH
 
 | | MicroED structure of mVDAC | | Descriptor: | Voltage-dependent anion-selective channel protein 1 | | Authors: | Martynowycz, M.W, Khan, F, Hattne, J, Abramson, J, Gonen, T. | | Deposit date: | 2020-11-25 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.12 Å) | | Cite: | MicroED structure of lipid-embedded mammalian mitochondrial voltage-dependent anion channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
