8AFZ
 
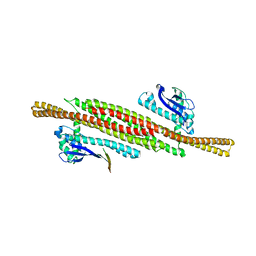 | | Architecture of the ESCPE-1 membrane coat | | Descriptor: | Cation-independent mannose-6-phosphate receptor, Sorting nexin-1, Sorting nexin-5 | | Authors: | Lopez-Robles, C, Scaramuzza, S, Astorga-Simon, E, Ishida, M, Williamsom, C.D, Banos-Mateos, S, Gil-Carton, D, Romero, M, Vidaurrazaga, A, Fernandez-Recio, J, Rojas, A.L, Bonifacino, J.S, Castano-Diez, D, Hierro, A. | | Deposit date: | 2022-07-18 | | Release date: | 2023-06-21 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Architecture of the ESCPE-1 membrane coat.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6SHN
 
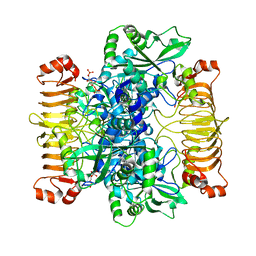 | | Escherichia coli AGPase in complex with FBP. Symmetry C1 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, Glucose-1-phosphate adenylyltransferase | | Authors: | Cifuente, J.O, Comino, N, D'Angelo, C, Marina, A, Gil-Carton, D, Albesa-Jove, D, Guerin, M.E. | | Deposit date: | 2019-08-07 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The allosteric control mechanism of bacterial glycogen biosynthesis disclosed by cryoEM.
Curr Res Struct Biol, 2, 2020
|
|
6SHJ
 
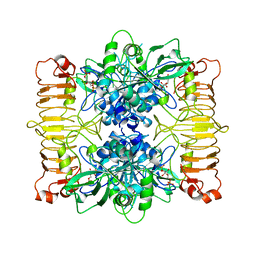 | | Escherichia coli AGPase in complex with FBP. Symmetry applied C2 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, Glucose-1-phosphate adenylyltransferase | | Authors: | Cifuente, J.O, Comino, N, D'Angelo, C, Marina, A, Gil-Carton, D, Albesa-Jove, D, Guerin, M.E. | | Deposit date: | 2019-08-07 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The allosteric control mechanism of bacterial glycogen biosynthesis disclosed by cryoEM.
Curr Res Struct Biol, 2, 2020
|
|
6SI8
 
 | | Escherichia coli AGPase in complex with AMP. | | Descriptor: | ADENOSINE MONOPHOSPHATE, Glucose-1-phosphate adenylyltransferase | | Authors: | Cifuente, J.O, Comino, N, D'Angelo, C, Marina, A, Gil-Carton, D, Albesa-Jove, D, Guerin, M.E. | | Deposit date: | 2019-08-09 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The allosteric control mechanism of bacterial glycogen biosynthesis disclosed by cryoEM.
Curr Res Struct Biol, 2, 2020
|
|
6SHQ
 
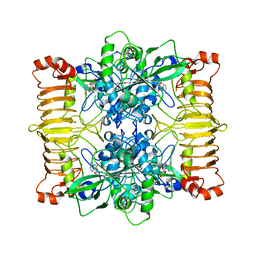 | | Escherichia coli AGPase in complex with AMP. Symmetry C2 | | Descriptor: | ADENOSINE MONOPHOSPHATE, Glucose-1-phosphate adenylyltransferase | | Authors: | Cifuente, J.O, Comino, N, D'Angelo, C, Marina, A, Gil-Carton, D, Albesa-Jove, D, Guerin, M.E. | | Deposit date: | 2019-08-07 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The allosteric control mechanism of bacterial glycogen biosynthesis disclosed by cryoEM.
Curr Res Struct Biol, 2, 2020
|
|
6H67
 
 | | Yeast RNA polymerase I elongation complex stalled by cyclobutane pyrimidine dimer (CPD) | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Sanz-Murillo, M, Xu, J, Gil-Carton, D, Wang, D, Fernandez-Tornero, C. | | Deposit date: | 2018-07-26 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of RNA polymerase I stalling at UV light-induced DNA damage.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6H68
 
 | | Yeast RNA polymerase I elongation complex stalled by cyclobutane pyrimidine dimer (CPD) with fully-ordered A49 | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Sanz-Murillo, M, Xu, J, Gil-Carton, D, Wang, D, Fernandez-Tornero, C. | | Deposit date: | 2018-07-26 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis of RNA polymerase I stalling at UV light-induced DNA damage.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7NWW
 
 | | CspA-27 cotranslational folding intermediate 1 | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Samatova, E, Macher, M, Liutkute, M, Gil-Carton, D, Novacek, J, Valle, M, Rodnina, M.V. | | Deposit date: | 2021-03-17 | | Release date: | 2022-01-19 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | A switch from alpha-helical to beta-strand conformation during co-translational protein folding.
Embo J., 41, 2022
|
|
7OIG
 
 | | CspA-27 cotranslational folding intermediate 3 | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Samatova, E, Macher, M, Liutkute, M, Gil-Carton, D, Novacek, J, Valle, M, Rodnina, M.V. | | Deposit date: | 2021-05-11 | | Release date: | 2022-01-19 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A switch from alpha-helical to beta-strand conformation during co-translational protein folding.
Embo J., 41, 2022
|
|
7OII
 
 | | CspA-70 cotranslational folding intermediate 2 | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Samatova, E, Macher, M, Liutkute, M, Gil-Carton, D, Novacek, J, Valle, M, Rodnina, M.V. | | Deposit date: | 2021-05-11 | | Release date: | 2022-01-19 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A switch from alpha-helical to beta-strand conformation during co-translational protein folding.
Embo J., 41, 2022
|
|
7OIF
 
 | | CspA-27 cotranslational folding intermediate 2 | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Samatova, E, Macher, M, Liutkute, M, Gil-Carton, D, Novacek, J, Valle, M, Rodnina, M.V. | | Deposit date: | 2021-05-11 | | Release date: | 2022-01-19 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A switch from alpha-helical to beta-strand conformation during co-translational protein folding.
Embo J., 41, 2022
|
|
7OT5
 
 | | CspA-70 cotranslational folding intermediate 1 | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Samatova, E, Macher, M, Liutkute, M, Gil-Carton, D, Novacek, J, Valle, M, Rodnina, M.V. | | Deposit date: | 2021-06-09 | | Release date: | 2022-01-19 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | A switch from alpha-helical to beta-strand conformation during co-translational protein folding.
Embo J., 41, 2022
|
|
5ME0
 
 | | Structure of the 30S Pre-Initiation Complex 1 (30S IC-1) Stalled by GE81112 | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Lopez-Alonso, J.P, Fabbretti, A, Kaminishi, T, Iturrioz, I, Brandi, L, Gil Carton, D, Gualerzi, C, Fucini, P, Connell, S. | | Deposit date: | 2016-11-14 | | Release date: | 2017-01-11 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (13.5 Å) | | Cite: | Structure of a 30S pre-initiation complex stalled by GE81112 reveals structural parallels in bacterial and eukaryotic protein synthesis initiation pathways.
Nucleic Acids Res., 45, 2017
|
|
5ME1
 
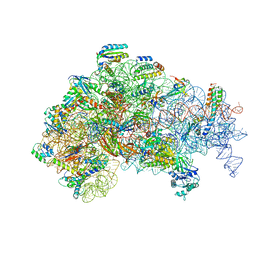 | | Structure of the 30S Pre-Initiation Complex 2 (30S IC-2) Stalled by GE81112 | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Lopez-Alonso, J.P, Fabbretti, A, Kaminishi, T, Iturrioz, I, Brandi, L, Gil Carton, D, Gualerzi, C, Fucini, P, Connell, S. | | Deposit date: | 2016-11-14 | | Release date: | 2017-01-11 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (13.5 Å) | | Cite: | Structure of a 30S pre-initiation complex stalled by GE81112 reveals structural parallels in bacterial and eukaryotic protein synthesis initiation pathways.
Nucleic Acids Res., 45, 2017
|
|
6R8B
 
 | | Escherichia coli AGPase in complex with FBP. | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, Glucose-1-phosphate adenylyltransferase | | Authors: | Cifuente, J.O, Comino, N, D'Angelo, C, Marina, A, Gil-Carton, D, Albesa-Jove, D, Guerin, M.E. | | Deposit date: | 2019-04-01 | | Release date: | 2020-02-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The allosteric control mechanism of bacterial glycogen biosynthesis disclosed by cryoEM
Biorxiv, 2020
|
|
6R8U
 
 | | Escherichia coli AGPase in complex with AMP. | | Descriptor: | ADENOSINE MONOPHOSPHATE, Glucose-1-phosphate adenylyltransferase | | Authors: | Cifuente, J.O, Comino, N, D'Angelo, C, Marina, A, Gil-Carton, D, Albesa-Jove, D, Guerin, M.E. | | Deposit date: | 2019-04-02 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The allosteric control mechanism of bacterial glycogen biosynthesis disclosed by cryoEM
Biorxiv, 2020
|
|
5LMX
 
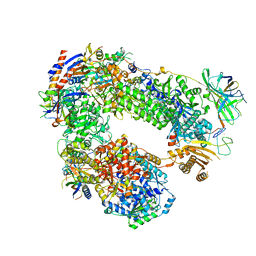 | | Monomeric RNA polymerase I at 4.9 A resolution | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Torreira, E, Louro, J.A, Gil-Carton, D, Gallego, O, Calvo, O, Fernandez-Tornero, C. | | Deposit date: | 2016-08-02 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | The dynamic assembly of distinct RNA polymerase I complexes modulates rDNA transcription.
Elife, 6, 2017
|
|
7NAS
 
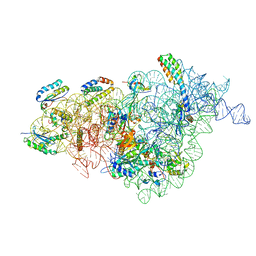 | | Bacterial 30S ribosomal subunit assembly complex state A (multibody refinement for body domain of 30S ribosome) | | Descriptor: | 16S rRNA, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-07-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
7NAU
 
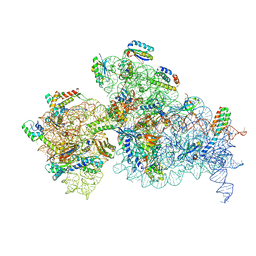 | | Bacterial 30S ribosomal subunit assembly complex state C (Consensus Refinement) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
7NAT
 
 | | Bacterial 30S ribosomal subunit assembly complex state A (Consensus refinement) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
7NAV
 
 | | Bacterial 30S ribosomal subunit assembly complex state D (Consensus refinement) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
7NAX
 
 | | Complete Bacterial 30S ribosomal subunit assembly complex state I (Consensus Refinement) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Lopez-Alonso, J, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
7NAR
 
 | | Complete Bacterial 30S ribosomal subunit assembly complex state F (+RsgA)(Consensus Refinement) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
7BOI
 
 | | Bacterial 30S ribosomal subunit assembly complex state F (multibody refinement for body domain of 30S ribosome) | | Descriptor: | 16S rRNA, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-07-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
7BOF
 
 | | Bacterial 30S ribosomal subunit assembly complex state I (body domain) | | Descriptor: | 16S rRNA, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Lopez-Alonso, J, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-07-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
