1VIG
 
 | | NMR STUDY OF VIGILIN, REPEAT 6, 40 STRUCTURES | | Descriptor: | VIGILIN | | Authors: | Musco, G, Stier, G, Joseph, C, Morelli, M.A.C, Nilges, M, Gibson, T.J, Pastore, A. | | Deposit date: | 1995-11-29 | | Release date: | 1996-04-03 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure and stability of the KH domain: molecular insights into the fragile X syndrome.
Cell(Cambridge,Mass.), 85, 1996
|
|
1VIH
 
 | | NMR STUDY OF VIGILIN, REPEAT 6, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | VIGILIN | | Authors: | Musco, G, Stier, G, Joseph, C, Morelli, M.A.C, Nilges, M, Gibson, T.J, Pastore, A. | | Deposit date: | 1995-11-29 | | Release date: | 1996-04-03 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure and stability of the KH domain: molecular insights into the fragile X syndrome.
Cell(Cambridge,Mass.), 85, 1996
|
|
1H5P
 
 | | Solution structure of the human Sp100b SAND domain by heteronuclear NMR. | | Descriptor: | NUCLEAR AUTOANTIGEN SP100-B | | Authors: | Bottomley, M.J, Liu, Z, Collard, M.W, Huggenvik, J.I, Gibson, T.J, Sattler, M. | | Deposit date: | 2001-05-24 | | Release date: | 2001-07-06 | | Last modified: | 2018-03-28 | | Method: | SOLUTION NMR | | Cite: | The SAND domain structure defines a novel DNA-binding fold in transcriptional regulation.
Nat. Struct. Biol., 8, 2001
|
|
2FMR
 
 | | KH1 FROM THE FRAGILE X PROTEIN FMR1, NMR, 18 STRUCTURES | | Descriptor: | FMR1 PROTEIN | | Authors: | Musco, G, Kharrat, A, Stier, G, Fraternali, F, Gibson, T.J, Nilges, M, Pastore, A. | | Deposit date: | 1997-10-06 | | Release date: | 1998-04-08 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the first KH domain of FMR1, the protein responsible for the fragile X syndrome.
Nat.Struct.Biol., 4, 1997
|
|
5NEW
 
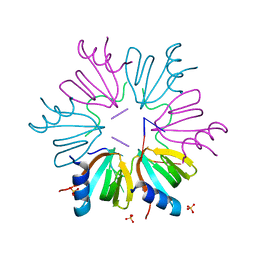 | |
6HIP
 
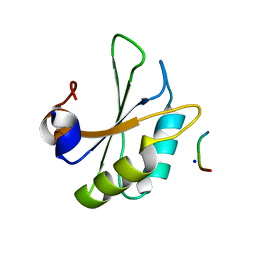 | | Structure of SPF45 UHM bound to HIV-1 Rev ULM | | Descriptor: | HIV-1 Rev (41-49), SODIUM ION, Splicing factor 45, ... | | Authors: | Pabis, M, Corsini, L, Sattler, M. | | Deposit date: | 2018-08-30 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Modulation of HIV-1 gene expression by binding of a ULM motif in the Rev protein to UHM-containing splicing factors.
Nucleic Acids Res., 47, 2019
|
|
7SMF
 
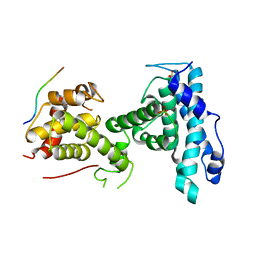 | | p107 pocket domain complexed with mutated HDAC1-3X peptide | | Descriptor: | Histone deacetylase 1, Retinoblastoma-like protein 1, SULFATE ION | | Authors: | Putta, S, Fernandez, S.M, Tripathi, S.M, Muller, G.A, Rubin, S.M. | | Deposit date: | 2021-10-25 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for tunable affinity and specificity of LxCxE-dependent protein interactions with the retinoblastoma protein family.
Structure, 30, 2022
|
|
7SME
 
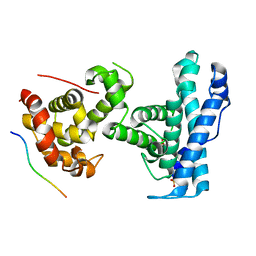 | | p107 pocket domain complexed with HDAC1 peptide | | Descriptor: | Histone deacetylase 1, Retinoblastoma-like protein 1, SULFATE ION | | Authors: | Putta, S, Fernandez, S.M, Tripathi, S.M, Muller, G.A, Rubin, S.M. | | Deposit date: | 2021-10-25 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural basis for tunable affinity and specificity of LxCxE-dependent protein interactions with the retinoblastoma protein family.
Structure, 30, 2022
|
|
7SMC
 
 | | p107 pocket domain complexed with ARID4A peptide | | Descriptor: | AT-rich interactive domain-containing protein 4A, Retinoblastoma-like protein 1, SULFATE ION | | Authors: | Putta, S, Fernandez, S.M, Tripathi, S.M, Muller, G.A, Rubin, S.M. | | Deposit date: | 2021-10-25 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for tunable affinity and specificity of LxCxE-dependent protein interactions with the retinoblastoma protein family.
Structure, 30, 2022
|
|
7SMD
 
 | | p107 pocket domain complexed with EID1 peptide | | Descriptor: | EP300-interacting inhibitor of differentiation 1, Retinoblastoma-like protein 1, SULFATE ION | | Authors: | Putta, S, Fernandez, S.M, Tripathi, S.M, Muller, G.A, Rubin, S.M. | | Deposit date: | 2021-10-25 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for tunable affinity and specificity of LxCxE-dependent protein interactions with the retinoblastoma protein family.
Structure, 30, 2022
|
|
3DXB
 
 | | Structure of the UHM domain of Puf60 fused to thioredoxin | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, thioredoxin N-terminally fused to Puf60(UHM) | | Authors: | Corsini, L, Hothorn, M, Scheffzek, K, Stier, G, Sattler, M. | | Deposit date: | 2008-07-24 | | Release date: | 2008-10-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dimerization and Protein Binding Specificity of the U2AF Homology Motif of the Splicing Factor Puf60.
J.Biol.Chem., 284, 2009
|
|
2JWP
 
 | | Malectin | | Descriptor: | Malectin | | Authors: | Schallus, T, Muhle-goll, C. | | Deposit date: | 2007-10-23 | | Release date: | 2008-08-12 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Malectin: A Novel Carbohydrate-binding Protein of the Endoplasmic Reticulum and a Candidate Player in the Early Steps of Protein N-Glycosylation
Mol.Cell.Biol., 19, 2008
|
|
2K46
 
 | | Xenopus laevis malectin complexed with nigerose (Glcalpha1-3Glc) | | Descriptor: | MGC80075 protein, alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose | | Authors: | Schallus, T. | | Deposit date: | 2008-05-28 | | Release date: | 2008-08-12 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Malectin: A Novel Carbohydrate-binding Protein of the Endoplasmic Reticulum and a Candidate Player in the Early Steps of Protein N-Glycosylation
Mol.Cell.Biol., 19, 2008
|
|
1LY7
 
 | | The solution structure of the the c-terminal domain of frataxin, the protein responsible for friedreich ataxia | | Descriptor: | frataxin | | Authors: | Musco, G, Stier, G, Kolmerer, B, Adinolfi, S, Martin, S, Frenkiel, T, Gibson, T, Pastore, A. | | Deposit date: | 2002-06-07 | | Release date: | 2002-06-26 | | Last modified: | 2022-12-21 | | Method: | SOLUTION NMR | | Cite: | Towards a structural understanding of Friedreich's
ataxia: the solution structure of frataxin
Structure Fold.Des., 8, 2000
|
|
